|
Home page Term 1 Term 2 Term 3 Term 4 About me Faculty website |
Domain evolutionThe LANC_like family (PF05147) was chosen, which is a lanthionine synthetase domain.Pfam ID: LANC_like Pfam AC: PF05147 Function: lanthionine bridge synthesis. Link to PFAM page: click here List of all domain architectures (on the Pfam website): click here Protein sequence alignment: fasta file, project file The two subtaxa chosen are Actinobacteria and Firmicutes (the taxon is Bacteria). The two architectures chosen are the following: This architecture is single-domain and is abbreviated A1 (for Actinobacteria) and F1 (for Firmicutes) in the tree. This architecture is two-domain and is abbreviated A2 (for Actinobacteria) and F2 (for Firmicutes) in the tree. The second domain has no known function. Here is the Excel pivot table, which was obtained using vlookup and a number of self-written python scripts, for example this one. Some sheets in the table are purely technical. This is the alignment with only the chosen sequences, and this is the project file. Empty and non-informative columns and short fragments were deleted from the alignment. The alignment contains quite a lot of conservative regions which makes it credible. This is the tree produced by Maximum Likelyhood algorithm in MEGA: 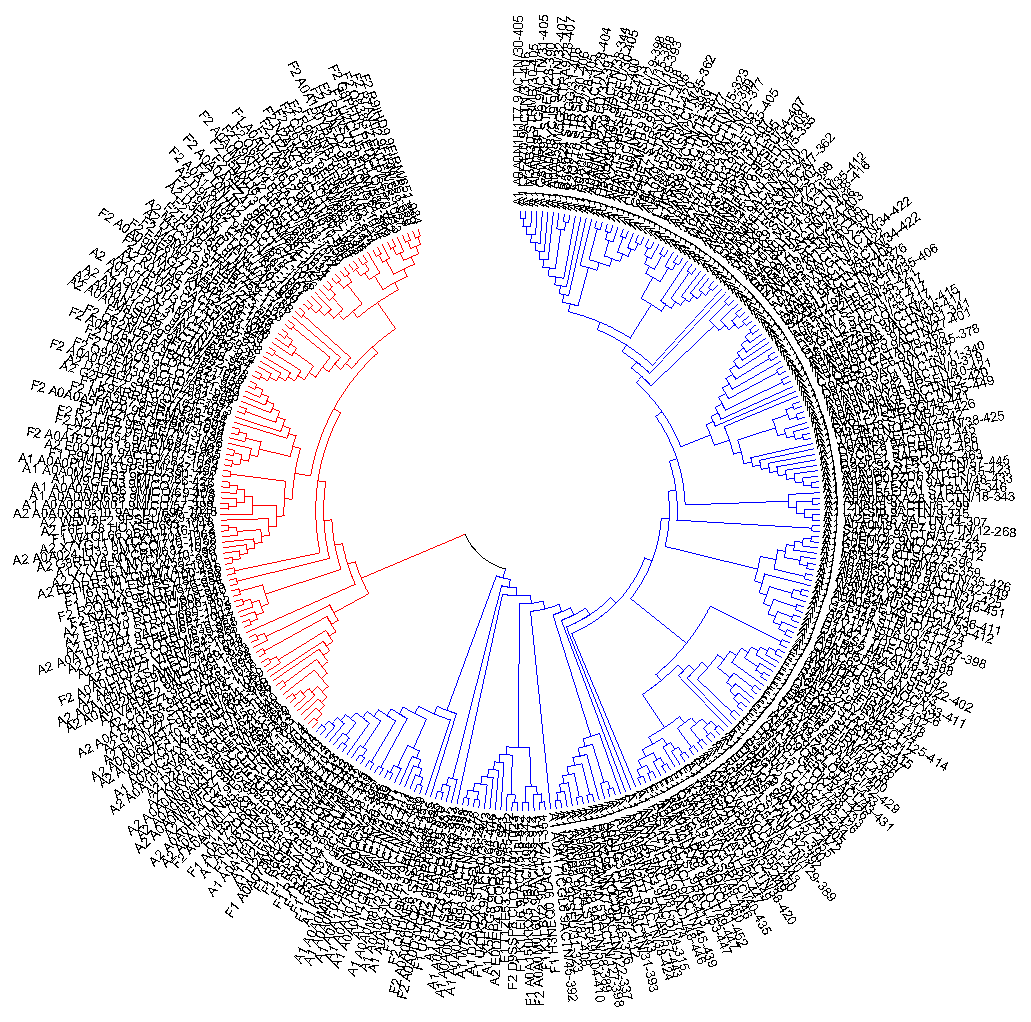 The red stands for the majority of proteins with the two-domain architecture, while the blue stands for the majority of the single-domain ones. You can find a magnified version here Tree Newick formula: NWK file As you can see, the division into the two architectures happened before Actinobacteria and Firmicutes biparted. Occasional occurence of single-domain architectures in the red section and of two-domain architecture in the right can be ascribed to mistakes in Pfam database. |
© Stanislav Tikhonov, 2018