|
Home page Term 1 Term 2 Term 3 Term 4 About me Faculty website |
Tree of homologsDescriptionThe task was to construct the tree of homologs of the protein CLPX_ECOLI (an ATP-dependent Clp protease ATP-binding subunit) among the proteins of the following bacteria:
For this task, standalone blastp was used. The TreeAfter aligning the blastp hits via muscle, the result was analyzed in Mega. The tree, which was reconstructed using the UPGMA algorithm, is presented below: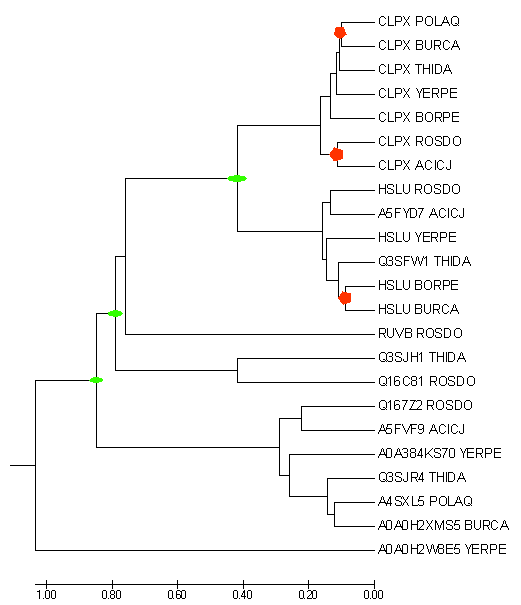
The AnalysisThe red shapes show places in the tree where species divergence has taken place (and orthologs were formed), and the green ellipses represent gene duplication events (formation of paralogs). Consequently, the pairs CLPX_POLAQ and CLPX_BURCA, CLPX_ROSDO and CLPX_ACICJ, HSLU_BORPE and HSLU_BURCA are examples of orthologs; the pairs CLPX_ROSDO and HSLU_ROSDO, Q3SJH1_THIDA and CLPX_THIDA, A4SXL5_POLAQ and CLPX_POLAQ are examples of paralogs. |
© Stanislav Tikhonov, 2018