|
Home page Term 1 Term 2 Term 3 Term 4 About me Faculty website |
KEGG PathwayGeneral Information on Inositol Phosphate Metabolism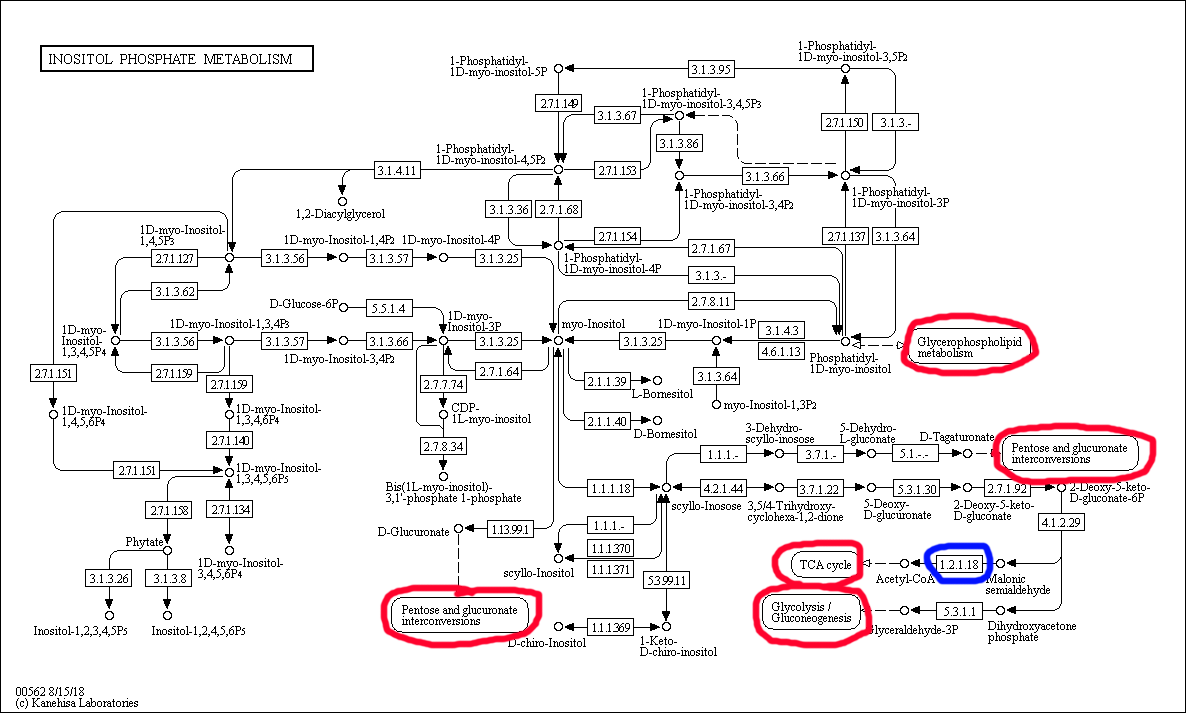 This map was obtained from the KEGG database, the indentifier is map00562: link Interconnections with other metabolic pathways are highlighted with red circles. The reaction chosen for the last task is highlighted in blue. Inositol comprises six-membered saturated ring with a hydroxyl group on each carbon atom, hence there are six sites for phosporylation in this molecule. Different phosphorylation derivatives of inositol participate in regulation of calcium channels, chromatin remodeling, and signalling (they can function as second messengers) in animals, phosphorus storage in plants (hexaphosphate), and assembly and maturation of HIV. In mammals, inositol phosphates either ingested or, according to the map, synthesized from phosphatidylinositol phosphates, which in their turn have been implicated in a plethora of various regulatory pathways and are membrane components (glycophospholipids). The products of inositol phosphate metabolism are acetyl CoA, glyceraldehyde phosphate, and different carbohydrates (and their phosphates). Inositol Phosphate metabolism in different domains of lifeThe three selected organisms are presented below:
 The enzymes the organism has are highlighted in green. It is clear from the map that these enzymes can act sequentially on one substrate. The sequences of reactions they catalyze start from the starting materials from the general map and lead to almost all of the final products on the general map. Overall, the organism is able to carry out many reactions from the reference map, approximately 60-65%. Oxidation of Malonic Semialdehyde into CO2 and Acetyl-CoA in KEGG DatabaseThis reaction was highlighted in blue on the reference map and has the indentifier R00705: linkReaction scheme: 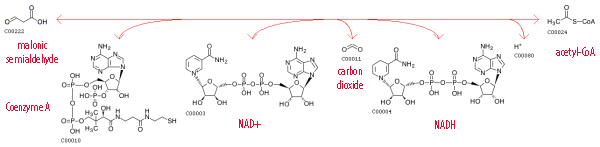
|
© Stanislav Tikhonov, 2018



