|
Home page Term 1 Term 2 Term 3 Term 4 About me Faculty website |
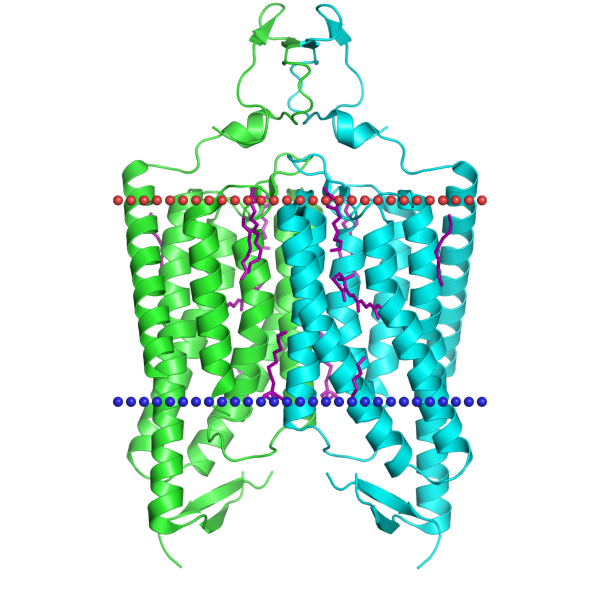
Transmembrane proteinsOPM databaseFor this task a channelrhodopsin was chosen (PDB ID: 3UG9), since channelrhodopsins are cool and are used in optogenetics by Ed Boyden at MIT (the most attentive readers might notice that this might be Michigan Institute of Technology, but no, you are wrong, go read your frigging books).Below a table describing some of its properties is presented:
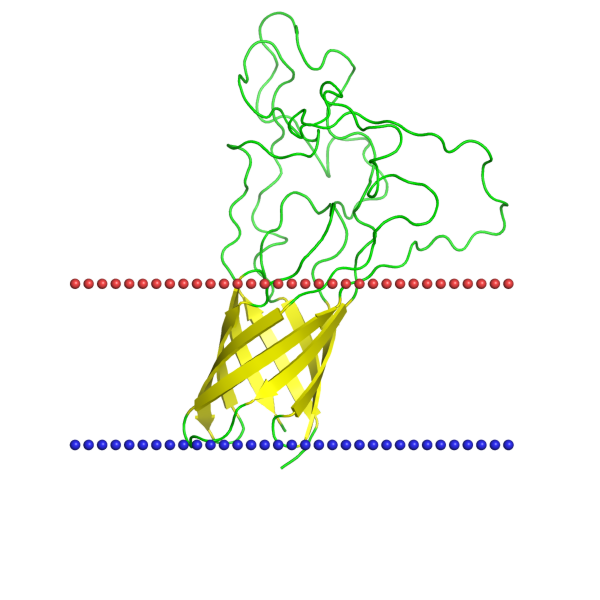
The protein of choice containing a β-barrel was an opacity rhodopsin (PDB ID: 2MAF). Honestly, I chose it because it has a cool name. Below you can find some of its properties:
Transmembrane segment predictionTwo web-services were used for prediction based on FASTA sequence: TMHMM and Phobius. Both predictors were run for the channelrhodopsin from the previous task.Results from TMHMM: # 3UG9:A|PDBID|CHAIN|SEQUENCE Length: 333 # 3UG9:A|PDBID|CHAIN|SEQUENCE Number of predicted TMHs: 5 # 3UG9:A|PDBID|CHAIN|SEQUENCE Exp number of AAs in TMHs: 96.86798 # 3UG9:A|PDBID|CHAIN|SEQUENCE Exp number, first 60 AAs: 0.24368 # 3UG9:A|PDBID|CHAIN|SEQUENCE Total prob of N-in: 0.46389 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 inside 1 65 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 TMhelix 66 88 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 outside 89 131 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 TMhelix 132 154 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 inside 155 166 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 TMhelix 167 184 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 outside 185 187 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 TMhelix 188 207 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 inside 208 226 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 TMhelix 227 249 3UG9:A|PDBID|CHAIN|SEQUENCE TMHMM2.0 outside 250 333 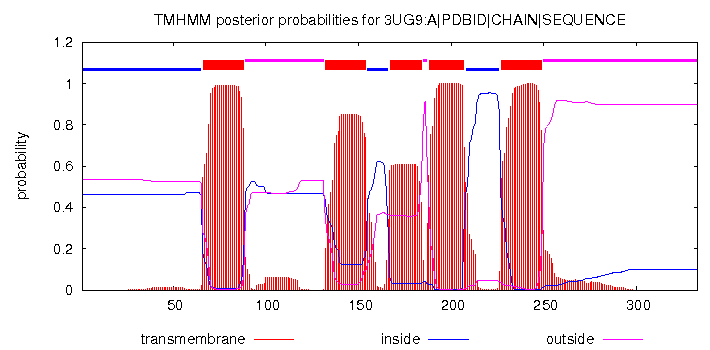 TMHMM absolutely accurately predicted helix No. 4, but failed to predict the coordinates of the others. After some careful inspection of the results, I noticed that it actually correctly predicted 5 out of 7 helices, but their coordinates are shifted towards the N-end by approximately 23 amino acids relative to the structure from the OPM database. The helices that it failed to predict are No. 2 and No. 7. The shift could be due to the fact that although there are 333 amino acids in the FASTA sequence, there are actually around 360 amino acids in the 3D structure on the PDB website. The ends are largely unstructured, so their position in space is by far not fixed, and they are a difficult target for the NMR machine. As a probable consequence, they were not shown in the FASTA sequence, which caused the shift in the coordinates. Results from TMHMM: ID 3UG9:A|PDBID|CHAIN|SEQUENCE FT TOPO_DOM 1 65 CYTOPLASMIC. FT TRANSMEM 66 88 FT TOPO_DOM 89 133 NON CYTOPLASMIC. FT TRANSMEM 134 154 FT TOPO_DOM 155 165 CYTOPLASMIC. FT TRANSMEM 166 185 FT TOPO_DOM 186 190 NON CYTOPLASMIC. FT TRANSMEM 191 209 FT TOPO_DOM 210 229 CYTOPLASMIC. FT TRANSMEM 230 249 FT TOPO_DOM 250 333 NON CYTOPLASMIC. // 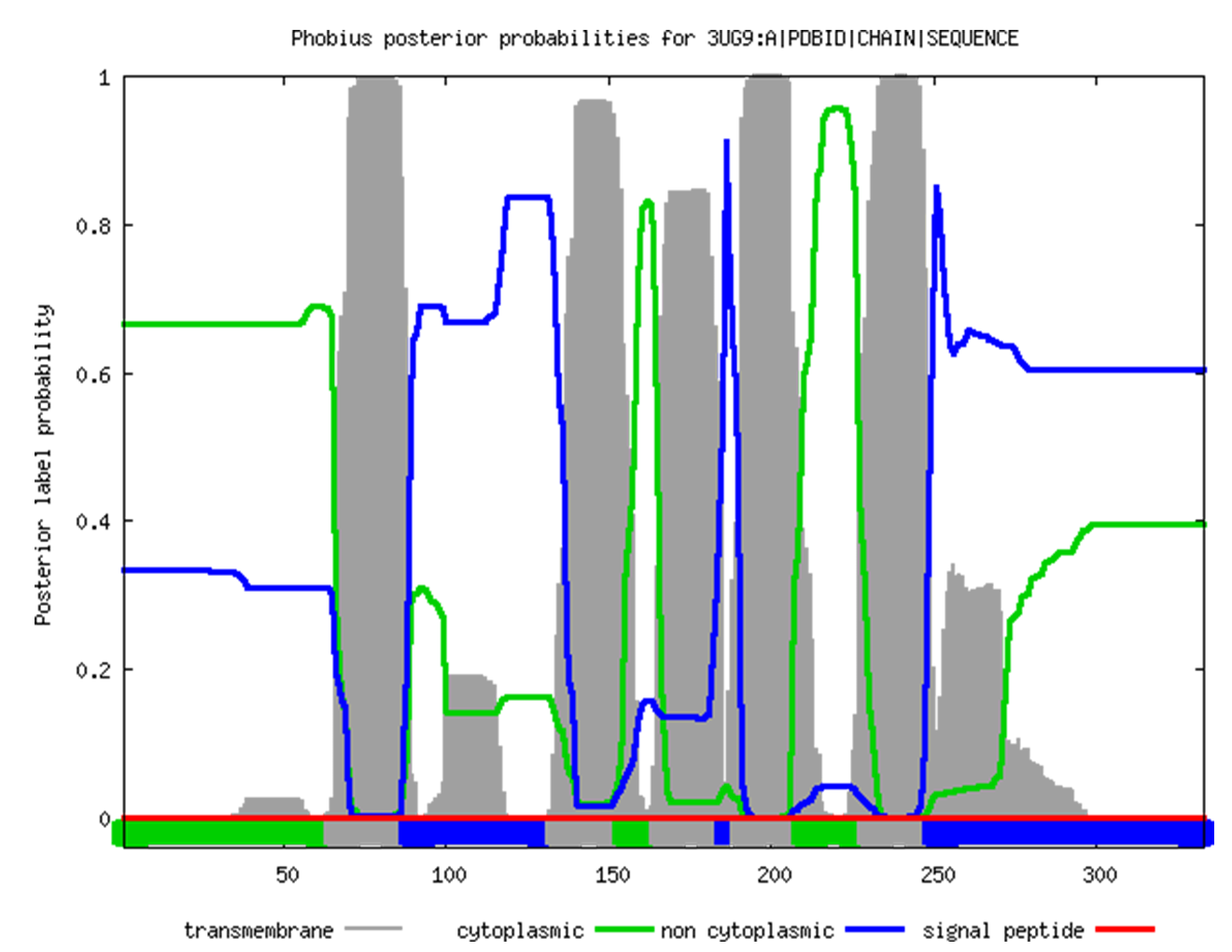 The results are almost the same as in the previous case, but Phobius did a better job at trying to find helices No. 2 and 7 (although didn't quite found them either). Interestingly, both websites failed to predict anything for the β-barrel protein, which can be either because they are taylored to be used with α-helices only, or because the 2MAF protein is located in the outer membrane of gram-negative bacteria, which is not polarized. It is possible that both factors affected the results. TCDBI beg your pardon, but since my website is in English, I decided to spare myself the burden of translating the TC-code into Russian. If needed, I will do it verbally, in person.Of the two proteins from this practical, only the channelrhodopsin turned out to be in TCDB, and its TCID is 3.E.1.7.2. It is broken down below: 3 - primary active transporters 3.E - light absorption-driven transporters 3.E.1 - ion-translocating microbial rhodopsin (MR) family The last two digits correspond to the subfamily and to the specific protein, respectively. |
© Stanislav Tikhonov, 2018