Defining the score cutoff
All the entries found by the HMM profile built (i.e. having E-value < 10) have been
gathered in the table2 and sorted by the score
decrease.
1291 proteins have been found, for each of which there are specificity and sensitivity of its generated
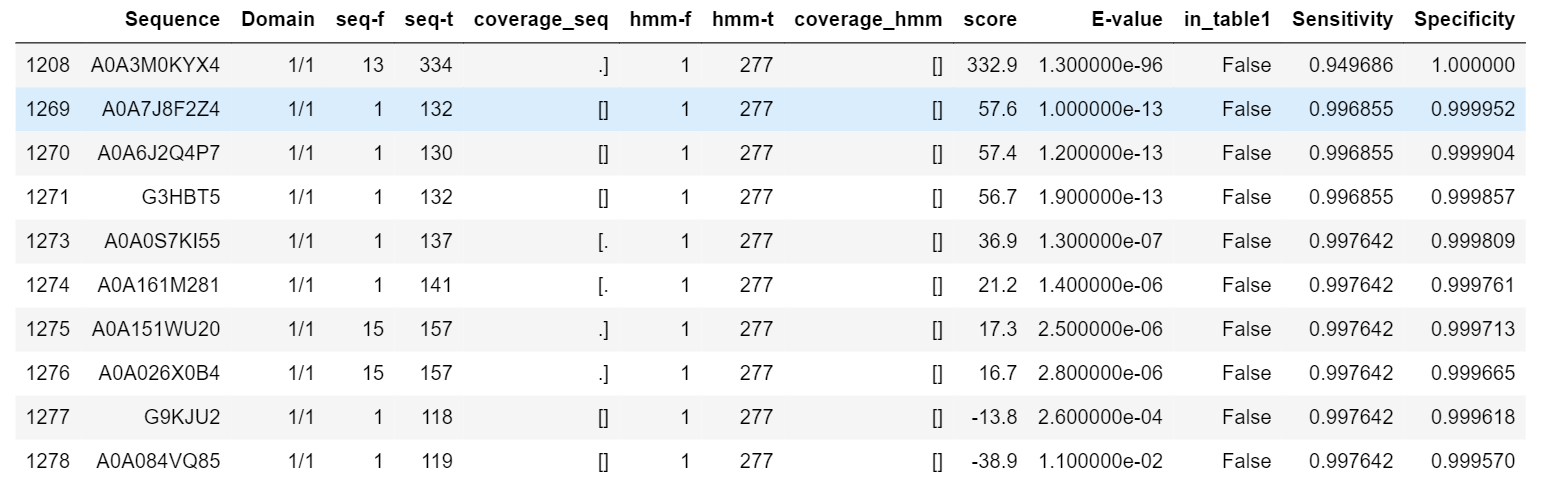
HMM search score as if it were taken as a cutoff. Specificity = (number of table_2 entries below the score cutoff and not in table_1) / (number of entries in table_1) Sensitivity = ((number of table_2 entries above the score cutoff and in table_1) + (number of entries rejected by HMM search)) / (number of entries with SOCS_box and without Ras) The first finding, absent from table1, (A0A3M0KYX4) has a distinctly high score and is followed by a number of findings with the target architecture (i.e. present in table1). According to InterPro annotation, A0A3M0KYX4 has Rab domain and SOCS-box domain; since Rab is a subfamily of Ras family and many proteins from table1 are annotated as somewhat like "Ras-related Rab protein", this protein may still be considered to have the target domain architecture. Pfam profile for Ras seems to be sensitive to the whole family, so it is quite unexpectedly to find the Ras domain unidentified by Pfam in this protein . The bulk of non-homologous findings starts, however, after the score falls below 58:
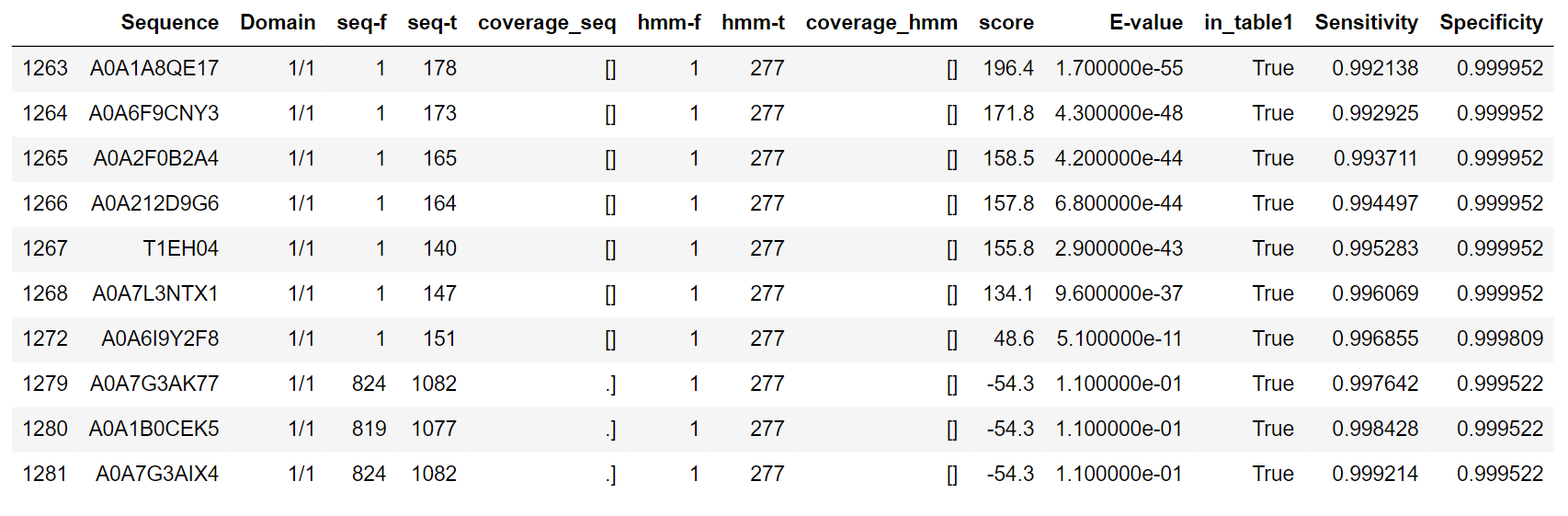
However, there are some target findings with score below 58. A0A6I9Y2F8 (score = 48.6) has been manually verified to have the target architecture in agreement with its placing in table1. Nevertheless, the next finding, A0A7G3AK77, (score = -54.3), though being present in table1, has inversed domain architecture (SOCS+Ras), so is not a target finding:
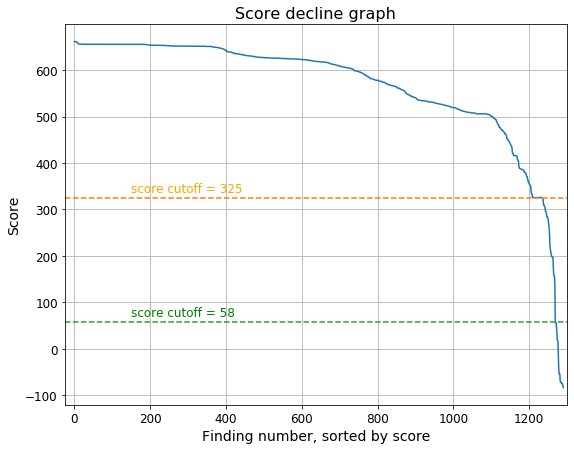
There is a steep score decrease after a rather common value of score = 325. However, there are a lot of target findings below this score and sequences with the mentioned stereotypic score = 325 all seem to belong to the large class of Aves (just numerous homologues):
So there could be defined the two cutoffs: a strict one (score > 325), based on the steep step on the Score decline graph, and a relaxed one (score > 58), based on the score of the second false finding. The relaxed cutoff seems better (more sensitive, than the strict one, though equally specific). |