In [1]:
from xmlrpclib import ServerProxy
In [2]:
import __main__
__main__.pymol_argv = [ 'pymol', '-x' ]
In [3]:
import pymol
pymol.finish_launching()
from pymol import cmd,stored
Изучаем возможности Pymol на примере лизоцима (1LMP):
In [4]:
cmd.fetch('1lmp')
Out[4]:
In [5]:
cmd.remove('solvent')
С помощью Sculpting можно вручную изменять структуру белка:
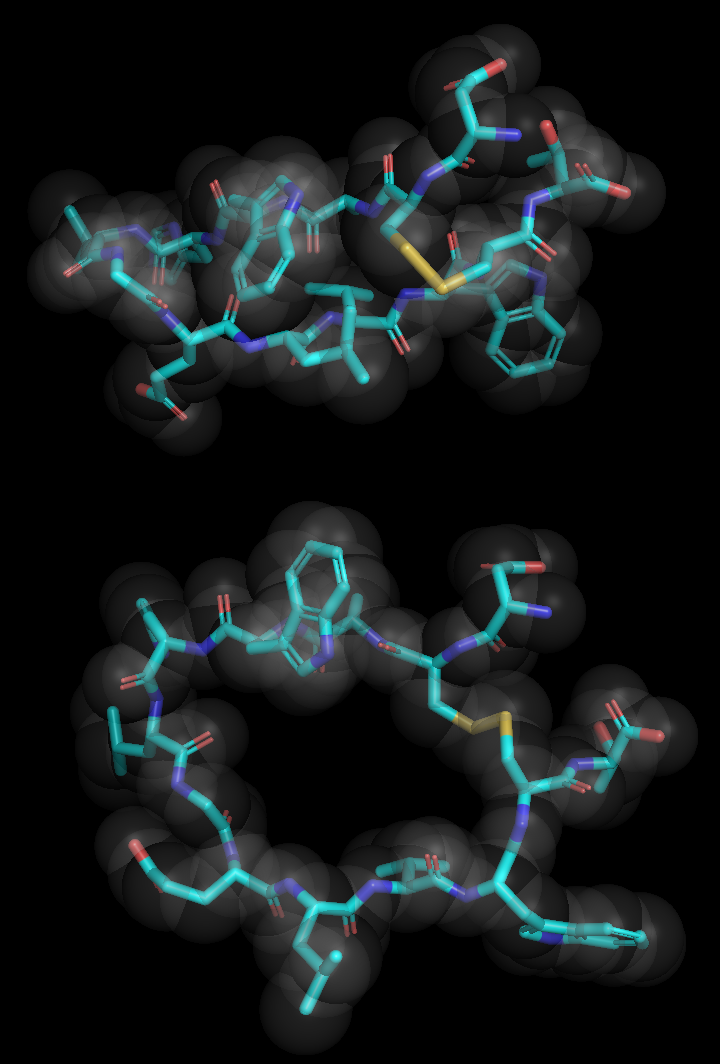
Сверху - исходная структура, снизу - измененная.
Мутация, которая приводит к потере связывания с лигандом:
In [6]:
cmd.reinitialize()
cmd.fetch('1lmp')
cmd.remove('solvent')
In [7]:
cmd.do('''
extract het, het
select env, byres /1lmp//A and (het around 3.5)
show lines, env
''')
In [8]:
cmd.do('''
distance hbonds, /1lmp//A, het, 3.2, mode=2
label env and name ca, resn,resi
''')
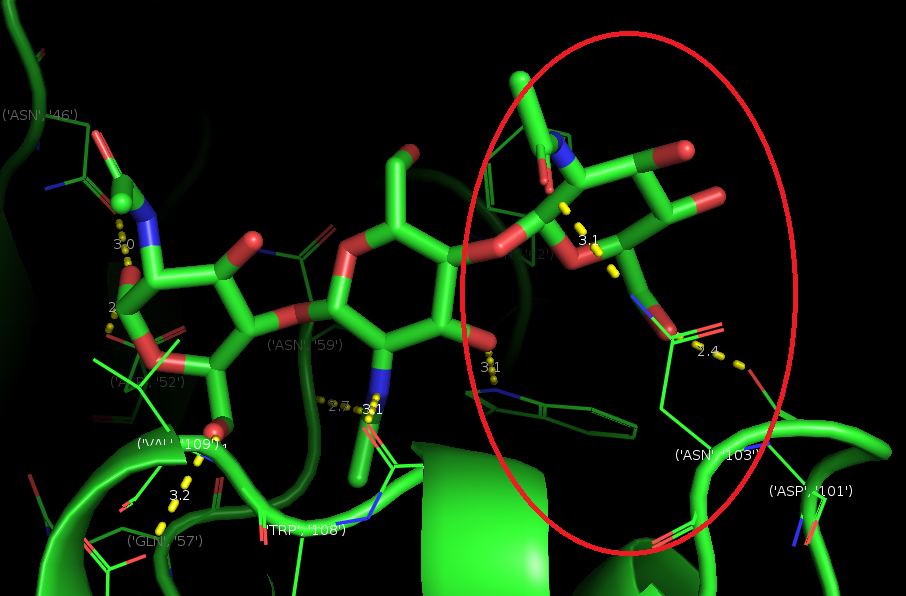
Лиганд и его окружение, в связывании с лигандом участвует аспарагин 103.
In [9]:
from pymol import wizard
In [10]:
cmd.wizard("mutagenesis")
cmd.do("refresh_wizard")
In [11]:
cmd.get_wizard().set_mode("ALA")
cmd.get_wizard().do_select("/1lmp//A/103")
cmd.get_wizard().apply()
cmd.set_wizard("done")
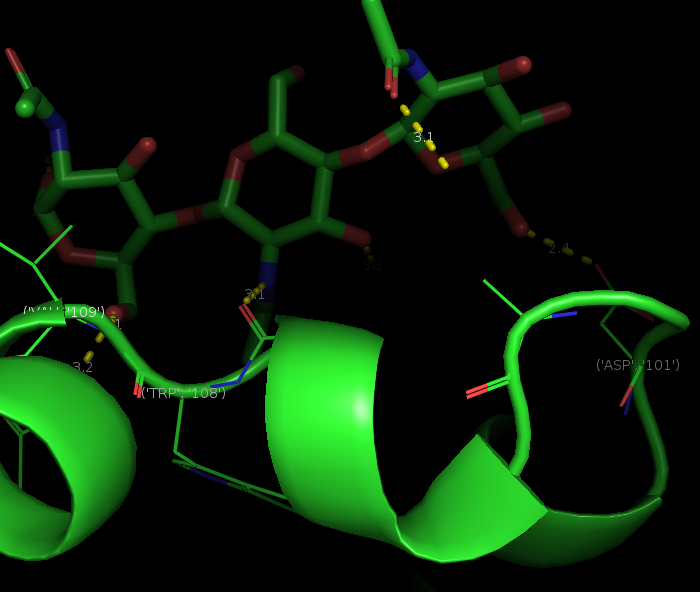
Лиганд и его окружение, участвующий в связывании с лигандом остаток заменен на аланин.
In [12]:
cmd.do('''
delete hbonds
hide lines
fetch 1lmp, 1lmp2
show sticks, het
show cartoons, 1lmp
show cartoons, 1lmp2
color yellow, 1lmp
color cyan, 1lmp2
color white, het
show sticks, /1lmp//A/ and resi 103
color orange, /1lmp//A/ and resi 103
show sticks, /1lmp2//A/ and resi 103
color blue, /1lmp2//A/ and resi 103
''')
In [13]:
cmd.remove('solvent')
In [14]:
cmd.do('''
set matrix_mode, 1
set movie_panel, 1
set cache_frames, 1
mset 1-150
frame 1
translate [0,60,0], object=1lmp2
mview store
mview store, object=1lmp2
mview store, object=1lmp
frame 60
translate [0,-60,0], object=1lmp2
mview store
mview store, object=1lmp2
mview store, object=1lmp
mview interpolate, object=1lmp2
frame 90
orient resi 103
mview store
mview store, object=1lmp2
mview store, object=1lmp
frame 120
orient resi 103
mview store
mview store, object=1lmp2
mview store, object=1lmp
''')
In [18]:
from IPython.display import HTML
HTML('<iframe src="https://player.vimeo.com/video/270170528" width="640" height="432" frameborder="0"></iframe>')
Out[18]:
In [16]:
cmd.reinitialize()
In [17]:
cmd.load('tamra.sdf')
Увы :(