Overview of the Klebsiella variicola genome and proteome
Co-responding Tumbinskii Roman
Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119234, Moscow, GSP-1, Leninskiye Gory, MSU, 1-73, Faculty of Bioengineering and Bioinformatics.
ABSTRACT
Motivation :
Klebsiella variicola is a human pathogen associated with high mortality in cases of bloodstream infection and diseases caused by K. pneumoniae. Moreover, there are instances of multidrug-resistant clinical isolates in such developed countries as the EU, the USA, and China. The better we understand the exact localization and mechanisms of resistance development the more likely we can arrange the correct treatment strategy to diminish the mortality of K. variicola species.KEYWORDS :
Klebsiella variicola, second Chargaff’s rule, protein lengths, nucleotide frequencies, K. variicola, CDS distributions1 INTRODUCTIONS
Monica Rosenblueth and colleagues proposed K. variicola in 2004. Using total DNA-DNA hybridization, monophyly in the phylogenetic analysis of 6 genes and comparisons of phenotypic features they concluded genetical isolation from K. pneumoniae strains. Found in plants and infected people in hospitals, this species does not have enzyme for adonitol breaking down, but carries gene of nitrogen-fixing protein (Mónica Rosenblueth, Lucía Martínez, Jesús Silva, Esperanza Martínez-Romero, 2004; Chen M, Li Y, Li S, Tang L, Zheng J, An Q., 2016.).
Its presence in blood or cerebrospinal fluid of patients with bloodstream infection (BSI) causes the highest mortality among K. pneumoniae sensu lato (Maatallah M, Vading M, Kabir MH, et al., 2014).
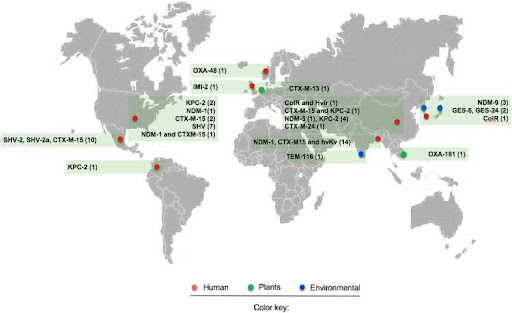
K. variicola carries a single chromosome and one plasmid. The proteome of this bacterium includes 5165 proteins encoded in one chromosome and 110 plasmid proteins. Previously, K. variicola species was resistant only to ampicillin, because of β-lactamase gene presence. This situation has become much more serious over time. Nowadays, as we can see in figure 3, there are 56 multidrug-resistant and hypervirulent isolates of K. variicola detected over the world and it seems that the environment plays a major role in accumulating antimicrobial-resistant genes (Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. 2019).
In this study, we analyzed the genome and proteome of the K. variicola, strain FH-1. The genome number and frequencies of nucleotides on one strand were calculated. As for proteome, the number of translating proteins, their lengths, and the number of molecules of different classes encoded in DNA were estimated.
2 METHODS
Both genome sequence and proteome features were downloaded from the Index of genomes server. Analysis of number and classes of genes encoded in DNA was computed with Google Sheets application, by creating a filter and inserting a column with numbers to calculate selected objects (Table 1). Protein lengths distribution was calculated and its chart was constructed with Google Sheets application by selecting column “length” and applying the function “Insert chart” (figures 1 and 2). For descriptive statistics estimations, such functions of Google Sheets application as “AVERAGE”, “STDEV”, “MEDIAN”, “min”, and “MAX” were applied to column “product_length”. Statistical significance of differences of CDS distributions was evaluated using a “BINOM.DIST” function of the Google Sheets application (Table 2). Nucleotides number was computed with the Python script.
3 RESULTS
3.1 Genome analysis
The overall size of the K. variicola genome is 5652418 bp. It consists only of 4 types of nucleotides: adenine (1205654 base pairs), thymine (1210478 base pairs), cytosine (1615382 base pairs), guanine (1620904 base pairs)). Frequencies for adenine and thymine on one strand are close in values (0.2133 and 0.2142) the same tendency is detected for guanine and cytosine frequencies (0.2858 and 0.2868).
These values were estimated for the chromosome and plasmid separately. One strand of the chromosome contains 1177849 adenine bases, 1182520 thymine bases, 1589038 guanine bases, and 1592863 cytosine bases. Frequencies for adenine and thymine are 0.2125 and 0.2134 and for guanine and cytosine are 0.2867 and 0.2874.
As for plasmid, its one strand carries 27805 adenine bases, 27958 thymine bases, 26344 guanine bases, and 28041 cytosine bases. Contents of adenine and thymine are 0.2524 and 0.2538, for guanine and cytosine frequencies are 0.2392 and 0.2546. The statistical evaluation shows that in almost all cases numbers of adenine and thymine bases as well as guanine and cytosine bases are not close. Frequencies of an equal amount of nucleotides for the whole genome are 0,001942 for adenine and thymine and 0,002140 for guanine and cytosine.
For the chromosome, the value for adenine and thymine is 0,002399 and for guanine and cytosine is 0,031962. For plasmid values are quite opposite: amounts of adenine and thymine are close, their frequency is 0,519783 and this result is statistically significant, but in the case of cytosine and guanine the result is 0.0000000000003.
3.2 Proteome and transcriptome analysis
Table 1 clarifies, that K. variicola chromosome consists of 2763 genes on its “+” strand and 2583 on its “-” strand. 2687 genes of chromosome “+” strand code proteins, 32 genes code tRNA molecules, and 34 genes are pseudogenes. As regards chromosomes “-” strand, 2478 genes are protein-coding, 54 genes code tRNA molecules and 24 are pseudogenes. 32 genes constitute “+” strand of plasmid and 86 genes - another one. However, only 1 gene on “+” strand and 7 genes on “-” strand are pseudogenes while the least are protein-coding genes. The plasmid sum of protein-coding genes and pseudogenes exactly equals the total number of genes for both strains.
Table 1.Number and class of genes encoded in DNA
| Type of DNA molecule | Number of genes | Number of proteins | Number of pseudogenes | Number of tRNA genes |
|---|---|---|---|---|
| Chromosome “+” strain | 2763 | 2687 | 34 | 32 |
| Chromosome “-” strain | 2583 | 2478 | 24 | 54 |
| Plasmid “+” strain | 32 | 31 | 1 | 0 |
| Plasmid “-” strain | 86 | 79 | 7 | 0 |
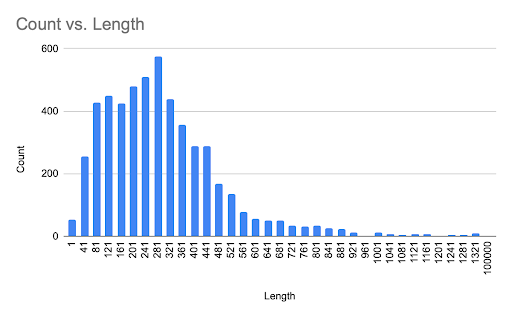
Analysis of protein lengths distribution, represented on figure 1, shows that the majority of proteins (575) encoded in the chromosome consisted of 281-321 amino acids. Another smaller pike represents proteins with lengths 81-121 amino acids. The only chromosome lacks any proteins with lengths 921-961 and 1161-1201 amino acids.
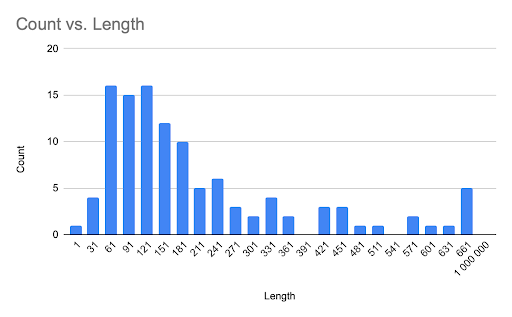
When it comes to plasmid, figure 2 is providing information that lengths 61-91 and 121-151 amino acids are the most widespread (16 proteins in both cases). However, there are four other smaller pikes equal to 211-241 amino acids, 301-331 amino acids, 391-421 and 421-451 amino acids. K. variicola's plasmid does not carry any proteins with lengths 361-391 and 511-541 amino acids.
Statistics on table 2 says that the average length of proteins is 312 amino acids, the standard deviation is 204,0 amino acids and the median equals 283 amino acids. Carbohydrate binding domain-containing protein encoded in the chromosome and has the greatest length among the whole proteome - 4196 amino acids. In contrast, pheST operon leader peptide PheM is the most short protein which length is equal to 14 amino acids. Both proteins are encoded in the chromosome.
Table 2. Descriptive statistics for proteins length
| Function type | Value | Protein name |
|---|---|---|
| Average length | 312 | — |
| Standard deviation | 204,0 | — |
| Median length | 283 | — |
| Maximum length | 4196 | carbohydrate binding domain-containing protein |
| Minimum length | 14 | pheST operon leader peptide PheM |
Coding DNA sequences distributed non arbitrary on “+” and “-” strands of chromosome (probability is less than 0,004), moreover, probability much less in cases of coding DNA sequences distribution on halves of each strand of the chromosome. For the “+” strand it is less than 0,00000000009 and less than 0,0000002 for the “-” strand.
4 DISCUSSION
From the side of genome analysis, the chromosome has a greater length and consists of more nucleotides than the plasmid. Statistical estimations let us conclude that the amount of adenine bases is close to the number of thymine bases only in the case of the plasmid. Hypothesis of equal numbers of adenine and thymine bases and cytosine and guanine bases is incorrect in cases of the whole genome of K. variicola, its chromosome and number of guanine and cytosine of the plasmid. It means that the second Chargaff’s rule is correct only in the case of adenine and thymine numbers of the plasmid.
Transcriptome and proteome analysis shows that the majority of proteins are encoded in the chromosome, moreover, it carries genes of proteins with greater lengths than the plasmid does. A remarkable feature is that the plasmid contains only protein-coding genes and pseudogenes while the chromosome additionally carries genes of tRNA, rRNA, ncRNA, RNase P RNA, tmRNA, antisense RNA, SRP RNA, and others. In addition, the chromosome carries genes of the smallest and the biggest proteins. Bacteria can use carbohydrate binding domain-containing proteins to recognize, bind and break down cellulose molecules or other soluble carbohydrates which can be found in plants (Ichikawa S, Karita S, Kondo M, Goto M., 2014). It makes sense since K.variicola can be found in plants. All these observations lead to the conclusion that the main reservoir of genetic information which also plays a key role in the regulation of the gene expression process of K.variicola is the chromosome, while the plasmid carries genes of supplementary and small proteins.
Supplementary materials
1) Python script for nucleotides number and frequency computation
2) Google Sheet table for statistical evaluation of nucleotides amount
3) FASTA file with listed CDS and proteins
4) Histograms and statistics significance for CDS distribution
ACKNOWLEDGEMENTS
We thank Alekseyevsky Andrey Vladimirovich, bioinformatics teacher, for providing themes and requirements for this study. We also would like to thank Rusinov Ivan Sergeevich, Spirin Sergey Alexandrovich and Arseniy Zinkevich for providing necessary knowledge and skills of Google Sheets, Terminal and Python script usage which were exceptionally useful during the research.
REFERENCES
Mónica Rosenblueth, Lucía Martínez, Jesús Silva, Esperanza Martínez-Romero, klebsiella variicola, A Novel Species with Clinical and Plant-Associated Isolates, Systematic and Applied Microbiology, Volume 27, Issue 1, 2004, Pages 27-35, ISSN 0723-2020, https://doi.org/10.1078/0723-2020-00261
Maatallah M, Vading M, Kabir MH, et al. klebsiella variicola is a frequent cause of bloodstream infection in the stockholm area, and associated with higher mortality compared to k. pneumoniae. PLoS One. 2014;9(11):e113539. Published 2014 Nov 26. doi:10.1371/journal.pone.0113539
Chen M, Li Y, Li S, Tang L, Zheng J, An Q. Genomic identification of nitrogen-fixing Klebsiella variicola, K. pneumoniae and K. quasipneumoniae. J Basic Microbiol. 2016 Jan;56(1):78-84. doi: 10.1002/jobm.201500415. Epub 2015 Oct 16. PMID: 26471769.
Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect. 2019;8(1):973-988. doi: 10.1080/22221751.2019.1634981. PMID: 31259664; PMCID: PMC6609320.
Ichikawa S, Karita S, Kondo M, Goto M. Cellulosomal carbohydrate-binding module from Clostridium josui binds to crystalline and non-crystalline cellulose, and soluble polysaccharides. FEBS Lett. 2014 Nov 3;588(21):3886-90. doi: 10.1016/j.febslet.2014.08.032. Epub 2014 Sep 11. PMID: 25217835.