Review
Arcobacter cloacae
Denis Urazov
Keywords: CDS; GC %; tRNA; tmRNA; ncRNA; rRNA; dnaE3|poIV
1. Introduction
Arcobacter cloacae (Taxonomic order - cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Arcobacteraceae; Arcobacter )[1] was first isolated from wastewater, but the first species of Arcobacter were found in domestic animals, present in both healthy and clinically ill pigs, being detected in 46% of samples of healthy swine. These spiral-shaped or slightly curved microbes are microaerophilic Gram-negative rods. Motility is by means of polar flagella, which may be either single or multiple, sheathed or naked. Arcobacter are cryophilic and oxygen tolerant species, they can grow and replicate in atmospheric oxygen and at temperatures about 15°С.[2] Most of the Arcobacter genus bacterias can cause human diseases, but such cases are rarely reported, yet those pathogens show their ability to be resistant to antibiotics and colonize food processing environments, which causes concern among scientists that Arcobacter can become serious problem and cause diseases in human population.
2. Materials and Methods
Review is based on provided genome features table (GCF_013201935.1_ASM1320193v1_feature_table.txt) and genomic sequence (GCF_013201935.1_ASM1320193v1_genomic.fna)[4]. All information found using Google Sheets and tables[4].
3. Results
3.1. Standard genome data
3.1.1. Amount of DNA, their identificational names and their length.
There were found 2 DNA chains: Chromosome and pACLO plasmid. The existence of plasmid proves that it is a prokaryotic organism since plasmids are often found in bacterias.
Table 1. (Data found using Google Sheets)
| Chromosome name | Type | Length (in pairs) |
| NZ_CP053833.1 | Chromosome | 2621653 |
| NZ_CP053834.1 | pACLO plasmid | 188833 |
As we can see (according to Table 1.), the chromosome has a much bigger length than plasmid. It can be explained by the specificity of plasmid. Plasmid doesn’t store any information that is vital for bacteria in normal conditions, but contains information that can be useful in harsh environments.
3.1.2 GC % in DNA
Table 2. (Data found using Google Sheets)
| Chromosome name | Type | GC |
| NZ_CP053833.1 | Chromosome | 27% |
| NZ_CP053834.1 | pACLO plasmid | 26% |
As we can observe (according to Table 2.), Arcobacter cloacae have low % of GC in its chromosome and plasmid. That means, this organism is related to low GC group (dnaE3|poIV)[5] , also resulting in low requirement of oxygen and low temperature latency (as was noted in introduction).[6]
3.2 Proteome proteins statistical data:
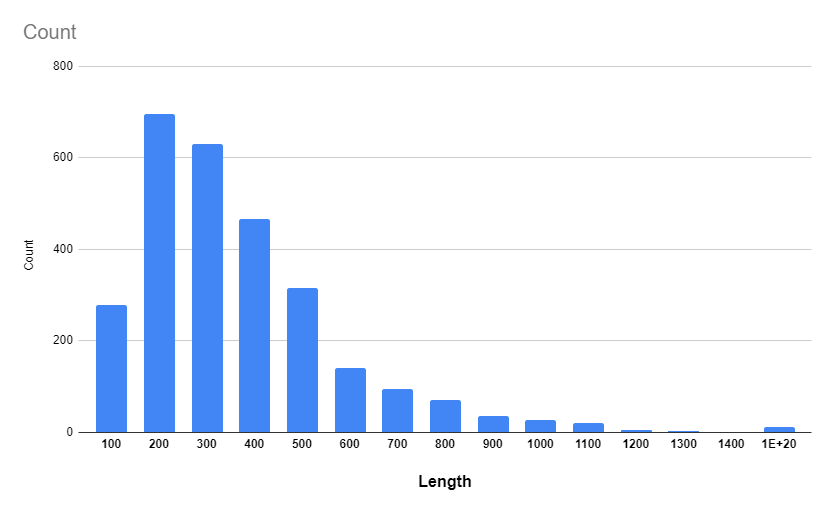
3.2.1 Protein lengths histogram.

According to the histogram (Figure 1.): proteins with length around 200 are the most common and proteins with length >= 1200 are incredibly rare. It is also worth noting that proteins with length ~100 occur in sequence only occasionally.
That means Arcobacter cloacae don’t produce proteins with extreme complexity or simplicity.
3.2.2 Compare the number of protein genes encoded on a direct chain and a complementary chain.
Table 3. (Data found using Google Sheets)
| DNA chain | Amount of protein genes | Total amount |
| Direct | 1282 | 2743 |
| Complementary | 1461 |
3.2.3 Find the amount of ribosomal proteins.
After analysis there were found 56 different ribosomal proteins. (The full list of ribosomal proteins found is attached: Proteins list)
Each of the ribosomal proteins is located on a chromosome, which is another proof of the specificity of the plasmid and that it doesn’t take part in normal vital activity of bacteria (does not produce ribosomal proteins, which are extremely important for life activity).
3.2.4 Reveal the number of hypothetical proteins and their percentage among the rest.
There are 490 hypothetical proteins. It’s around 17,86 % out of all proteins encrypted in the genome. That means that the Arcobacter Cloacae genome is quite well researched.
Table 4. (Data found using Google Sheets)
| Hypothetical proteins amount | Ratio comparing to amount of all proteins |
| 490 | 17,86% |
3.2.5 Find the amount of transporter proteins and their ratio to all proteins.
91 transporter proteins were found. It’s around 3,3 % out of all proteins encrypted in the genome. It is interesting that information about same proteins is stored in different parts of DNA (The full list of transporter proteins found is attached: Transporter proteins list)
Example of proteins in different parts of DNA, according to the attached table:
Table 5. (Data found using Google Sheets)
| Name | Start | End |
| ABC transporter ATP-binding protein | 662365 | 663048 |
| ABC transporter ATP-binding protein | 1961911 | 1962708 |
Same proteins can be synthesized on different parts of DNA. It allows Arcobacter Cloacae to produce them in various ways, so in case of mutation in one place Arcobacter will have a backup option to create the vital protein.
3.3 RNA statistical data.
3.3.1 Compare the amount of RNA genes and protein genes.
According to 3.2.2 there are 2743 protein genes. After further research, 76 RNA genes were found (tRNA, tmRNA, ncRNA, rRNA). As we can see, there are around 36x times more protein genes.
3.3.2 Find the amount of ribosomal RNA.
The amount of ribosomal RNA (rRNA) is equal 3 (with 16S ribosomal RNA, 5S ribosomal RNA and 23S ribosomal RNA repeating 5 times).
3.3.3 Find the amount of transportation RNA (tRNA).
There are 21 unique tRNAs (for Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, His, ile, Leu, Lys, Met, Phe, Pro, Sec, Ser, Thr, Trp, Tyl, Val transportation). Each one except for tRNA-Cys, tRNA-His, tRNA-Pro, tRNA-Sec and tRNA-Trp are encoded multiple times. Such interchangeability helps Arcobacter Cloacae to produce more tRNAs for use.
4. Intersecting genes:
Genes that share the same coordinates will be counted as intersecting.
4.1.1
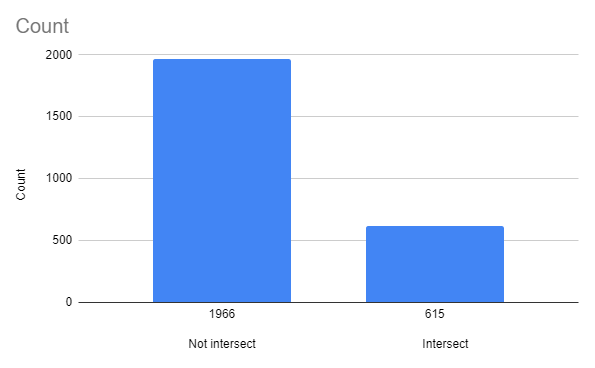
Table 6. Intersecting genes in the chromosome. (Data found with the usage of Google Sheets)
| Chromosome | Intersect | Starts where previous ends |
| Complete | 615 | 126 |
| Strand + | 252 | 53 |
| Strand - | 302 | 73 |
4.1.2 Intersection diagram:
Figure 3 shows an amount of intersecting genes compared to the amount of not intersecting genes.
4.2.1
Table 7. Intersecting genes in the plasmid. (Data found using Google Sheets)
| pACLO plasmid | Intersect | Starts where previous ends |
| Complete | 40 | 8 |
| Strand + | 21 | 6 |
| Strand - | 18 | 2 |
4.2.2 Intersection diagram:
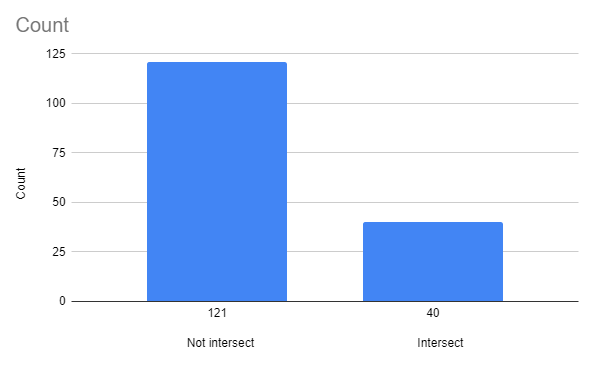
Figure 4 shows an amount of intersecting genes compared to the amount of not intersecting genes.
As we can see (Figure 3., Table 6. and Figure 4., Table 7.), most of the genes do not intersect, but if they do - there’s an approximately identical chance that such an event will occur in both strands (+ and -).
High amounts of intersecting genes may lead to a mutation. We can conclude that Arcobacter cloacae bacterias are exposed for mutations.
4. Conclusion:
Author of this microreview does not find Arcobacter Cloacae to be prospective in future research, yet this bacteria should be supervisioned to prevent it becoming dangerous to the human population.
5. References: