Natribaculum luteum(Halovarius luteus): mini-review
Veronika Dorofeeva
Faculty of Bioengineering and Bioinformatics, Moscow State University
Abstract
Natribaculum luteum is a poorly studied species from the class Halobacteria. This mini-review observes the characteristics and genome of this species.
Introduction
Natribaculum luteum is an archaeon from the genus Natribaculum, family Halobacteriaceae. Detailed taxonomy is provided in Table 1.
Table 1. Lineage of Natribaculum luteum.
| Superkingdom | Archaea |
| Kingdom | Methanobacteriati |
| Phylum | Methanobacteriota |
| Clade | Stenosarchaea |
| Class | Halobacteria |
| Order | Halobacteriales |
| Family | Natrialbaceae |
| Genus | Natribaculum |
| Current name | Natribaculum luteum |
The genus Natribaculum[1] was classified as a separate taxon only in 2015, so this archaeon has hardly been studied. More information is available on the Halobacteriaceae family since it was discovered in 1974. The active transport system in species of this family, including Natribaculum luteum, is of interest, since they inhabit only highly saline environments.
I analysed the genome and proteome of the archaea Natribaculum luteum to identify its genetic characteristics.
Materials and Methods
The taxonomy of the archaea was obtained from NCBI’s taxonomy browser.
Genomic and proteomic sequences were downloaded from the NCBI electronic database. The link is provided in supplementary materials.
PubMed was used as a source of scientific literature on archaea under study.
Google Sheets were used to pinpoint the difference between the length of Natribaculum luteum’s proteins as well as to analyse the intersections on + and - strands.
Blast was used as an alignment tool to build a tree for Natribaculum luteum’s bacteriorhodopsin.
PDB was a source of 3d models of bacteriorhodopsin and halorhodopsin.
Results
1.Histogram of proteins’ lengths
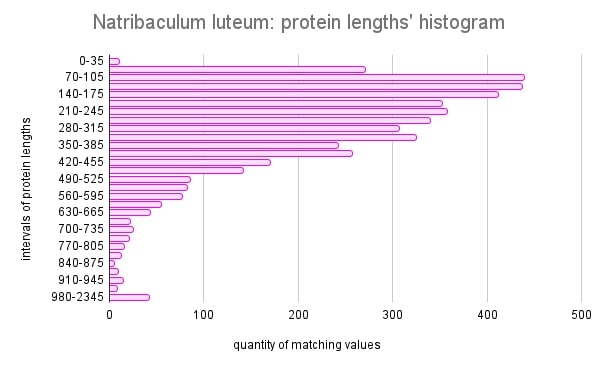
According to the histogram of protein lengths in Figure 1, the amino acid sequences of Natribaculum luteum are mostly short. The average length is 130 amino acid residues. This may be conditioned by the fact that this species is evolutionarily closer to bacteria[2]. It can be assumed that proteins with short sequences most often form simple structures, since there are few connections and interactions between amino acid residues. Bacterial proteins tend to be shorter on average, as eukaryotes have more complex cellular structures and systems, which requires longer and more diverse proteins to perform various functions.
Figure 1. Histogram of proteins’ lengths for Natribaculum luteum.
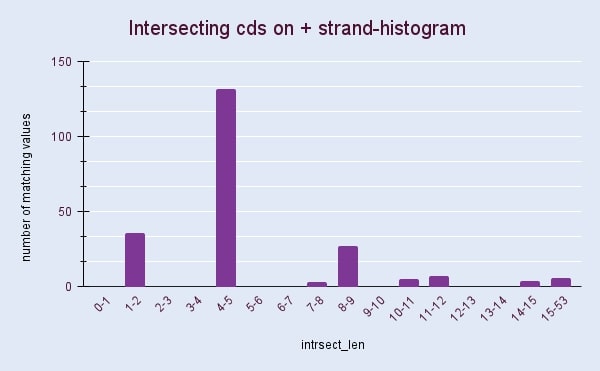
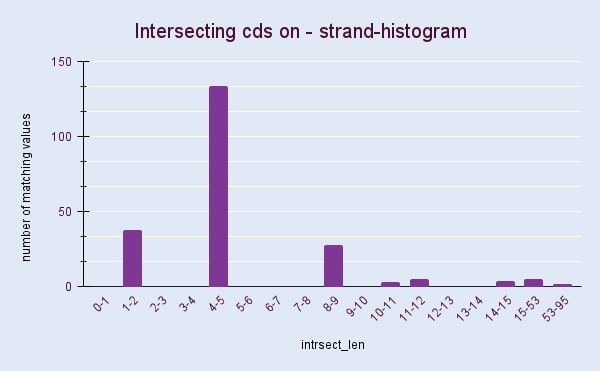
2. The lengths of intersections between coding sequences of + and - strands
Figures 2 and 3 show histograms of intersection lengths for Natribaculum luteum’s coding sequences. The asymmetry in the organization of genes[3] is usually a reason for the difference in the length and number of intersections on the + and – chains. However obtained histograms show surprisingly similar results. According to the figures calculated with Google Sheets, intersecting CDS account for 9.4 and 9.6 percent of the total number of sequences, respectively, for the + and – chain. Such intersections ensure the compactness of the genetic material, which is necessary in conditions when the cell is small.
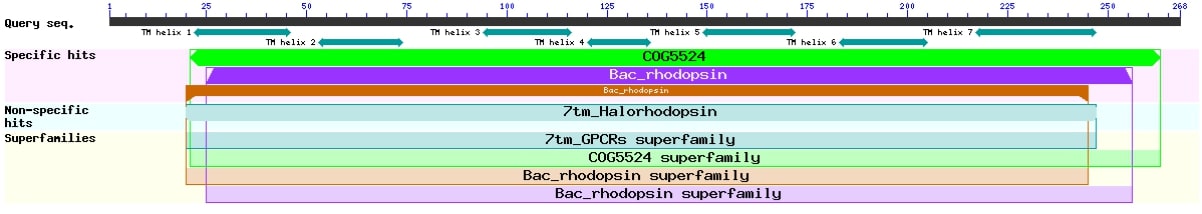
3. The distance tree for bacteriorhodopsin.
Although Natribaculum luteum isn’t well-studied, as a member of the Halobacteria class this species has certain features. For example its DNA contains a sequence encoding the bacteriorhodopsin protein[4], which classification is presented in Figure 4.
Figure 4. Graphical summary of conserved domains identified on the sequence of Natribaculum luteum’s bacteriorhodopsin.
This purple protein is of great interest, since it works as the proton pump, converting light energy into chemical energy. Its special structure allows this species to live in a hypersaline environment. Using the Blast tool for bacteriorhodopsin, the sequences closest in amino acid composition were found, the data obtained are shown in Figure 5.
Figure 5. Distance tree of closest alignments for Natribaculum luteum’s bacteriorhodopsin.
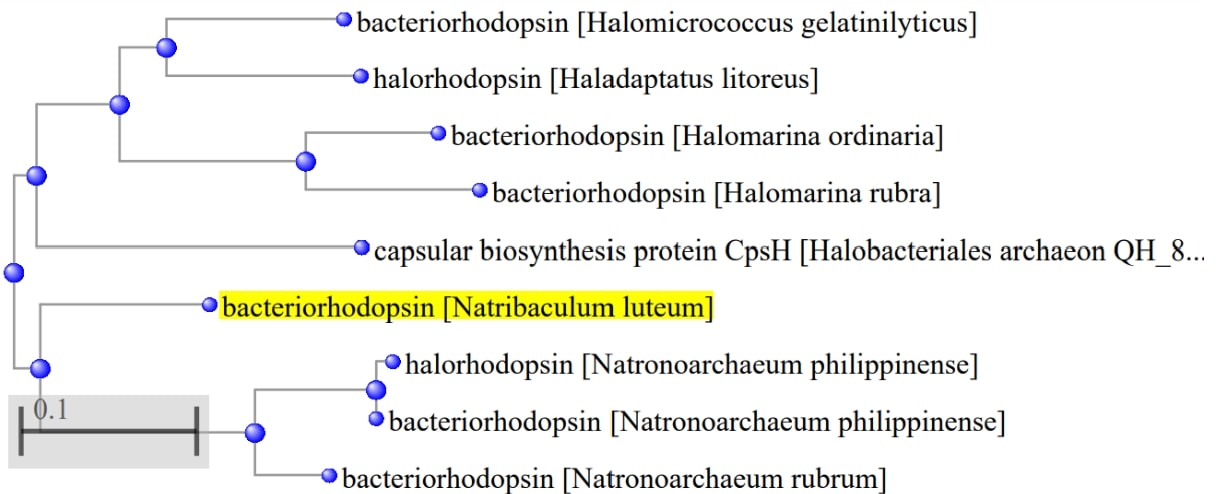
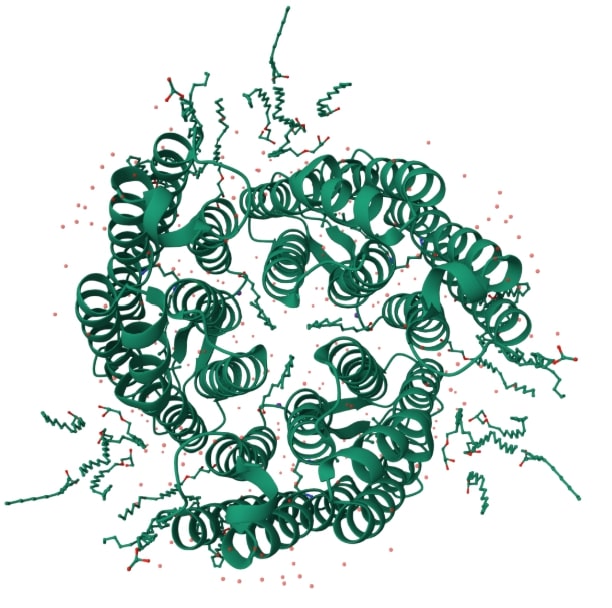
Based on the similarity of the amino acid composition, it can be concluded about the evolutionary proximity of the corresponding species found in China and the Philippines[5]. It is seen from the tree that bacteriorhodopsin is also similar in composition to halorhodopsin. The similarity of their primary configurations is reflected in the appearance of these proteins, as can be seen from Illustration 1 and 2.
Illustration 1. Structure of bacteriorhodopsin(6RQP)[6]
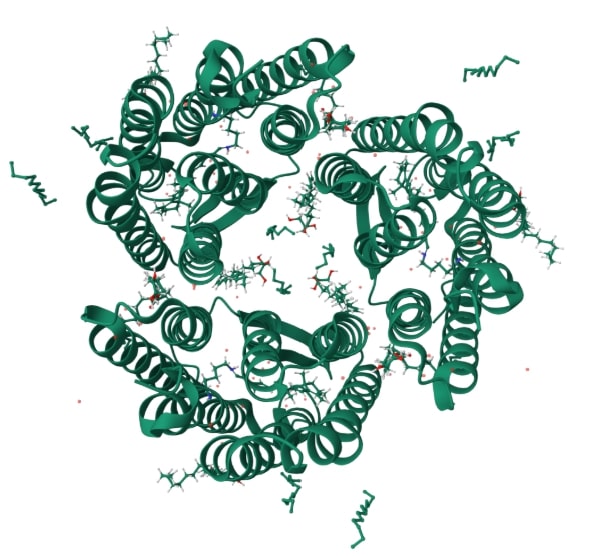
Illustration 2. Structure of halorhodopsin(1E12)[7]
References
[1]Sun YP, Wang BB, Zheng XW, Wu ZP, Hou J, Cui HL. Description of Halosolutus amylolyticus gen. nov., sp. nov., Halosolutus halophilus sp. nov. and Halosolutus gelatinilyticus sp. nov., and genome-based taxonomy of genera Natribaculum and Halovarius. Int J Syst Evol Microbiol. 2022 Oct;72(10). doi: 10.1099/ijsem.0.005598. PMID: 36288096.
[2]Woese CR, Magrum LJ, Fox GE. Archaebacteria. J Mol Evol. 1978 Aug 2;11(3):245-51. doi: 10.1007/BF01734485. PMID: 691075.
[3]Moeckel C, Zaravinos A, Georgakopoulos-Soares I. Strand asymmetries across genomic processes. Comput Struct Biotechnol J. 2023 Mar 11;21:2036-2047. doi: 10.1016/j.csbj.2023.03.007. PMID: 36968020; PMCID: PMC10030826.
[4]L016/s0005-2728(00)00124-9. PMID: 10984585.anyi JK. Bacteriorhodopsin. Biochim Biophys Acta. 2000 Aug 30;1460(1):1-3. doi: 10.1
[5]Shimane Y, Nagaoka S, Minegishi H, Kamekura M, Echigo A, Hatada Y, Ito T, Usami R. Natronoarchaeum philippinense sp. nov., a haloarchaeon isolated from commercial solar salt. Int J Syst Evol Microbiol. 2013 Mar;63(Pt 3):920-924. doi: 10.1099/ijs.0.042549-0. Epub 2012 Jun 1. PMID: 22659499.
[6]Tobias Weinert et al.,Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.Science365,61-65(2019).DOI:10.1126/science.aaw8634
[7]Michael Kolbe et al.,Structure of the Light-Driven Chloride Pump Halorhodopsin at 1.8 Å Resolution.Science288,1390-1396(2000).DOI:10.1126/science.288.5470.1390
Supplementary materials
Portal, from which genomic and proteomic sequences were borrowed:
https://docs.google.com/spreadsheets - histogram of protein lengths
https://docs.google.com/spreadsheets - histogram of intersecting CDS on + strand
https://docs.google.com/spreadsheets - histogram of intersecting CDS on - strand
Acknowledgments
To Ivan Sergeevish for setting off my bash test. It supported me so much while I was rewriting this review.
To my friend Dima, who helped with the original task.