Computing of pairwise sequence alignments
In this Jalview project 3 pairwise alignments of homologous proteins - ATPE_ADICA and ATPE_CLOBL - are presented. They are calculated by using 3 different programs: muscle, needle, and water (paired respectively)
The first difference
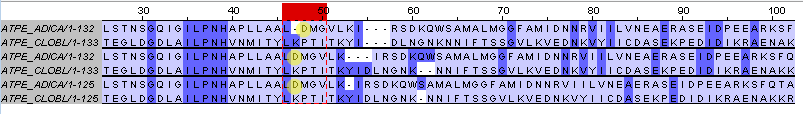
As It can be seen from the picture, in the first group of alignments, made by muscle program by deleting other sequences, ASP(45) is paired with PRO(48) while in other groups it is paired with LYS(47)
The second difference
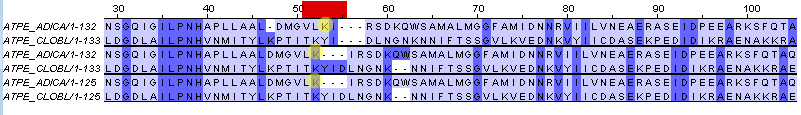
As It can be seen from this picture, in the second and the third groups of alignments, made by needle and water respectively, LYS(50) is paired with LYS(52) while in the first group it is paired with TYR(53)
The third difference

As It can be seen from this picture, in the second and the third groups of alignments LYS(55) is paired with LYS(60) while in the first group it is paired with GLY(58)
Comment
Considering that the first group is made by aligning it with other homologous sequences, I suggest that it is the best variant since it takes into consideration other residues.