Taxonomy and function
To perform the task, the sequence obtained during the execution of task 6 was taken. Then, the nucleotide BLAST with the blastn algorithm (several similar sequences) and Nucleotide database (nr / nt) with default parameters was used.

According to the results it can be concluded that this gene is mitochondrial gene encoding cytochrome C oxidase 1 subunit of the respiratory complex I.
JalView alignment with first 9 sequenses was made, proving the previous statement. Thus, since All 10 downloaded sequences downloaded belong to the same species, it can be said that the source of the selected sequences is the Ophiopholis aculeata species (Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Echinodermata; Eleutherozoa; Asterozoa; Ophiuroidea; Ophiuridea; Ophiurida; Ophiurina; Gnathophiurina; Ophiactidae; Ophiopholis).

Comparing the results of Different BLAST Algorithms
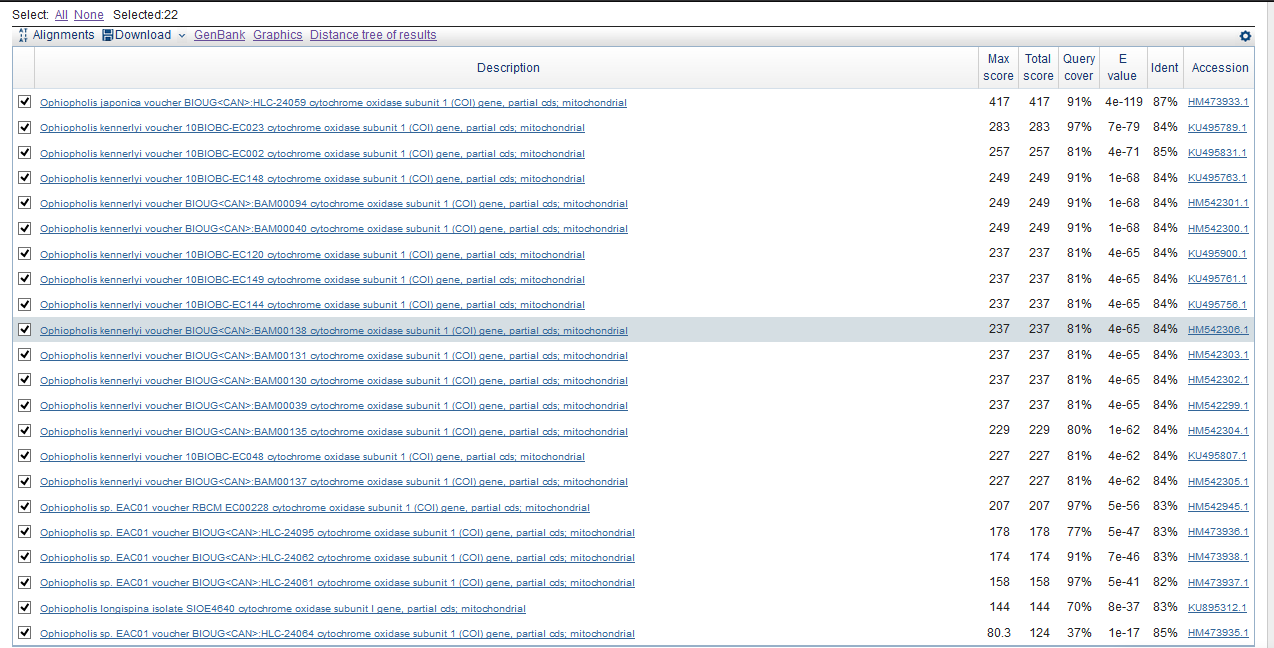
To compare them, you need to expand your search. Results of the query for the genus Ophiactidae (taxid: 41169): 15 sequences for megablast, 22 for default blasn, 22 for modified blastn.
| Algorithm | Database | Max Target Sequences | Expect Threshold | Word Size | Max matches | Match/Mismatch Scores | Gap Costs |
| megablast | Nucleotide collection (nr/nt) | 1000 | 0.001 | 28 | 0 | 1, -2 | Linear |
| blastn | Nucleotide collection (nr/nt) | 1000 | 0.001 | 11 | 0 | 2 -3 | Existence:5, Extention: 2 |
| blastn | Nucleotide collection (nr/nt) | 1000 | 0.001 | 7 | 0 | 1 -4 | Existence:5, Extention: 2 |


| Algorithm comparison | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Algorithm | The amount of findings | Best score | worst score | best E-value | worst E-value | best Ident | worst Ident | best Query cover | worst Query cover |
| megablast | 15 | 710 | 549 | 0.0 | 6e-159 | 86% | 82% | 98% | 99% |
| default blastn | 22 | 774 | 462 | 0.0 | 5e-132 | 86% | 81% | 98% | 73% |
| modified blastn | 22 | 417 | 80.3 | 4e-132 | 1e-17 | 87% | 85% | 91% | 37% |
According to the data we, can conclude that blastn and megablast find the same sequences, but differ in parameters max score, total score and query cover. Megablast works much more strictly, crossing out a greater number of finds and, therefore, giving out sequences that are only closest to the original. It is suitable for finding closely related sequences, it works quite quickly.
Homologous proteins
The task was performed using the BLAST+ version installed on kodomo. First, a local database (makeblastdb -in mybase.fasta -dbtype nucl) was created. Then, for each of the selected proteins tblastn algorithm that finds protein homologs in the formal translation of the nucleotide bank was used (tblastn -query **.fasta -db mybase > *.out)
HSP71_YEAST
May play a role in the transport of polypeptides both across the mitochondrial membranes and into the endoplasmic reticulum. The best find has a good E-value and other parameters, so that this find can be called a homologue, and most likely it has similar functions.
EIF3G_SCHPO

RNA-binding component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is involved in protein synthesis of a specialized repertoire of mRNAs and, together with other initiation factors, stimulates binding of mRNA and methionyl-tRNAi to the 40S ribosome. The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation. This subunit can bind 18S rRNA. The results are quite decent thus that can be called as a homologue
PRPC_EMENI

Catalyzes the synthesis of (2S,3S)-2-methylcitrate from propionyl-CoA and oxaloacetate and also from acetyl-CoA and oxaloacetate with a greater efficiency. Also has citrate synthase activity and can substitute for the loss of citA activity. The finding can be described as positive, however E-value is pretty low and thus there is no similar functions.
Protein gene in one of the contigs
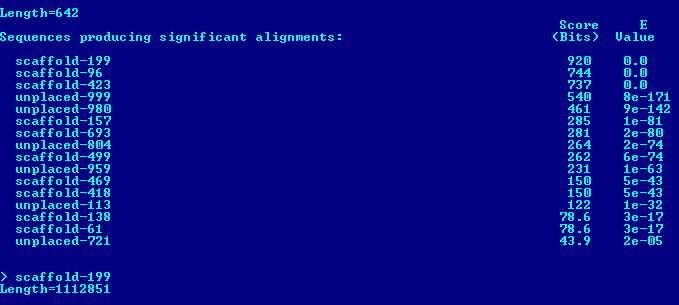
Sequence scaffold-26 was chosen as long enough to have a protein gene (483268 np). Then, megablast was launched with the default parameters and restrictions on the taxon Fungi (taxid:4751).
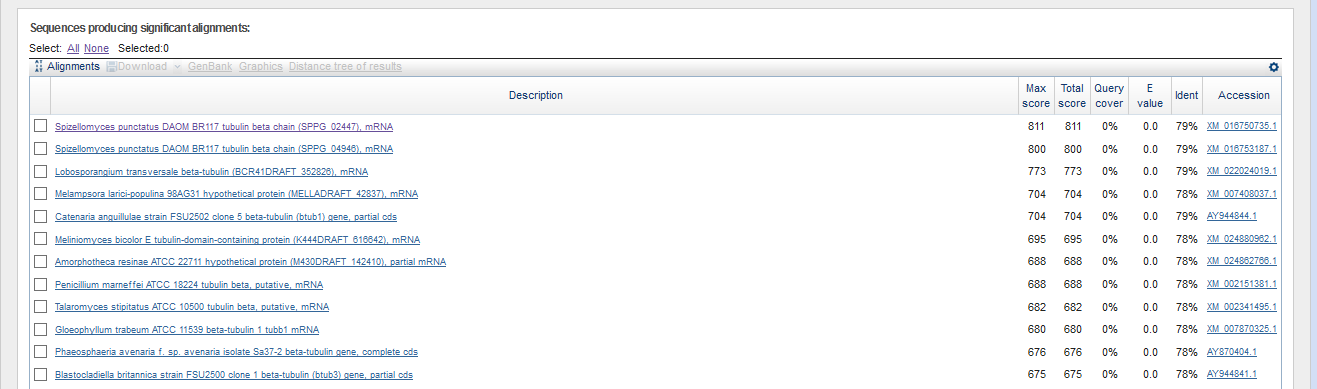
As you can see from the results, we have gene sequences with good Identity (79%) and E-value (0). Therefore, it is safe to say that this contig contains tubulin beta chain protein
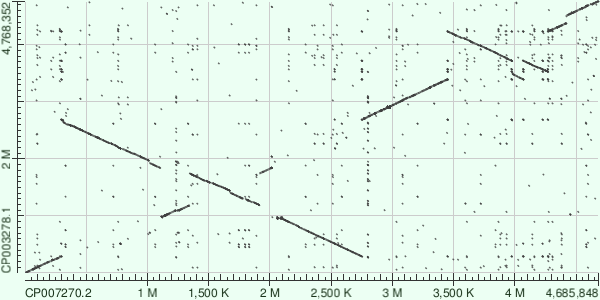
Dot matrix view
Salmonella enterica subsp. enterica serovar Enteritidis str. EC20121176 (NCBI Reference Sequence: CP007270.2) and Salmonella enterica subsp. enterica serovar Typhi str. P-stx-12 (NCBI Reference Sequence: CP003278.1) were chosen. Then, megablast with default parameters was used in order to make the matrix. All in all, it can be said that these two sequences are relatively similar, although, examples of inversions are clearly seen.