Command table
| Command | Description |
|---|---|
| cat /P/y16/term3/block3/adapters/*.fa >> adapters.fasta | File with adaptors |
| java -jar /nfs/srv/databases/ngs/suvorova/trimmomatic/trimmomatic-0.30.jar SE -phred33 SRR4240379.fastq SRR.fastq ILLUMINACLIP:adapters.fasta:2:7:7 java -jar /nfs/srv/databases/ngs/suvorova/trimmomatic/trimmomatic-0.30.jar SE -phred33 SRR.fastq SRRtrimm.fastq TRAILING:20 MINLEN:30 | Adapters deletion and sequence trimming After adapters deletion: Input Reads: 7400155 Surviving: 7269845 (98,24%) Dropped: 130310 (1,76%) After trimming: Input Reads: 7269845 Surviving: 6993284 (96,20%) Dropped: 276561 (3,80%) |
| velveth ./velvet 29 -short -fastq SRRtrimm.fastq | Velveth helps you construct the dataset for the following program, velvetg, and indicate to the system what each sequence file represents. 29 is the lendth of k-mers |
| velvetg ./ | Velvetg is the core of Velvet where the de Bruijn graph is built then manipulated. Results: n50 of 31053, max 82103 |
Contig analysis
| ID | Length (k-mers) | E-value | % Identity | Gaps / % | Chains | Chr coordinates | Part chr length | Read coordinates | Part read length | Coverage |
| 5 | 82103 | 0.0/td> | 77% | 2%/td> | +/+ | 451729 - 529004 | 77276 | 2388 - 82131 | 79744 | 47.938393 |
| 2 | 70497 | 0.0 | 81 | 2% | +/+ | 528977 - 594099 | 65423 | 1-67134 | 67134 | 49.610836 |
| 6 | 49941 | 0.0 | 75 | 4% | +/+ | 49941 - 173180 | 123240 | 53 - 45435 | 45383 | 48.604492 |

The longest contig №5, as it can be seen, there are 14 different alingments:
1. 451729 - 454069
2. 462496 - 467421
3. 467412 - 474667
4. 474844 - 480660
5. 480874 - 481545
6. 481997 - 488106
And 8 more...
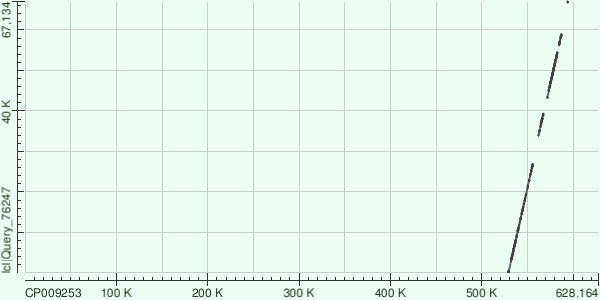
Number of Matches: 8. The gap between the last two alignments is big and therefore it can be presumed that this contig is not as similar to the original sequence as desired.
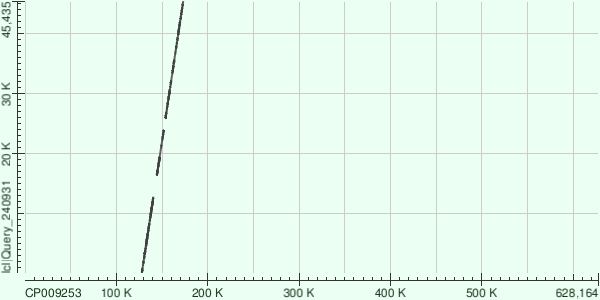
Number of Matches: 5.
1. 127825 - 140555
2. 144368 - 151796
3. 153752 - 161738
4. 161898 - 166752
5. 166750 - 173180
All in all, it is can be concluded that the final assemly is acceptable.