Сравнение значений торсионных углов.
Таблица по результатам измерений:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| A-форма |
-51.67 |
174.79 |
41.73 |
79.11 |
-147.75 |
-75.15 |
-157.18 |
| B-форма |
-29.89 |
136.32 |
31.17 |
143.34 |
-140.77 |
-160.50 |
-97.97 |
Значения торсионных углов из презентации:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| A-форма |
-62 |
173 |
52 |
88 или 3 |
178 |
-50 |
-160 |
| B-форма |
-63 |
171 |
54 |
123 или 131 |
155 |
-90 |
-117 |
A-форма.
Средние значения по анализу find_pair и analyze:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| A-форма |
-51.70 |
174.80 |
41.70 |
79.08 |
-147.7 |
-75.0 |
-157.2 |
Main chain and chi torsion angles:
Note: alpha: O3'(i-1)-P-O5'-C5'
beta: P-O5'-C5'-C4'
gamma: O5'-C5'-C4'-C3'
delta: C5'-C4'-C3'-O3'
epsilon: C4'-C3'-O3'-P(i+1)
zeta: C3'-O3'-P(i+1)-O5'(i+1)
chi for pyrimidines(Y): O4'-C1'-N1-C2
chi for purines(R): O4'-C1'-N9-C4
Strand I
base alpha beta gamma delta epsilon zeta chi
1 G --- 174.8 41.7 79.0 -147.8 -75.1 -157.2
2 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
3 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
4 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
5 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
6 A -51.7 174.8 41.7 79.0 -147.8 -75.1 -157.2
7 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
8 C -51.7 174.8 41.7 79.0 -147.8 -75.0 -157.2
9 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
10 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
11 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
12 C -51.7 174.8 41.7 79.1 -147.7 -75.1 -157.2
13 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
14 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
15 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
16 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
17 G -51.7 174.8 41.7 79.0 -147.8 -75.1 -157.2
18 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
19 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
20 C -51.7 174.8 41.7 79.1 --- --- -157.2
Strand II
base alpha beta gamma delta epsilon zeta chi
1 C -51.7 174.8 41.7 79.0 --- --- -157.2
2 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
3 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
4 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
5 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
6 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
7 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
8 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
9 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
10 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
11 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
12 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
13 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
14 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
15 A -51.7 174.8 41.7 79.0 -147.8 -75.1 -157.2
16 G -51.7 174.8 41.7 79.1 -147.7 -75.1 -157.2
17 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
18 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
19 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2
20 G --- 174.8 41.7 79.1 -147.8 -75.1 -157.2
B-форма.
Средние значения по анализу find_pair и analyze:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| B-форма |
-29.9 |
136.3 |
31.2 |
143.3 |
-140.8 |
-160.5 |
-98 |
Strand I
base alpha beta gamma delta epsilon zeta chi
1 G --- 136.4 31.1 143.4 -140.8 -160.5 -98.0
2 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
3 T -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9
4 C -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
5 G -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
6 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
7 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
8 C -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9
9 G -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
10 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
11 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
12 C -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
13 G -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0
14 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
15 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
16 C -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
17 G -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
18 A -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0
19 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
20 C -29.9 136.3 31.2 143.3 --- --- -98.0
Strand II
base alpha beta gamma delta epsilon zeta chi
1 C -29.9 136.4 31.1 143.4 --- --- -98.0
2 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
3 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
4 G -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0
5 C -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
6 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
7 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
8 G -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
9 C -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9
10 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
11 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
12 G -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
13 C -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
14 T -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9
15 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
16 G -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
17 C -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0
18 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0
19 A -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0
20 G --- 136.3 31.2 143.3 -140.8 -160.5 -98.0
Z-форма.
Средние значения по анализу find_pair и analyze:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| Z-форма |
-139.5 или 52 |
-136.7 или 179 |
-173.8 или 50.9 |
94.9 или 137.6 |
-100.4 |
-64.8 или 82 |
-154.7 или 58.7 |
Strand I
base alpha beta gamma delta epsilon zeta chi
1 G --- 179.0 -173.8 94.9 -103.6 -64.8 58.7
2 C -139.5 -136.7 50.9 137.6 -96.5 81.9 -154.3
3 G 52.0 179.0 -173.8 94.9 -103.6 -64.8 58.7
4 C -139.5 -136.8 50.8 137.6 -96.5 82.0 -154.3
5 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7
6 C -139.5 -136.8 50.9 137.6 -96.5 82.0 -154.3
7 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7
8 C -139.5 -136.7 50.9 137.6 -96.5 81.9 -154.3
9 G 52.0 179.0 -173.8 94.9 -103.6 -64.8 58.7
10 C -139.5 -136.8 50.8 137.6 --- --- -154.3
Strand II
base alpha beta gamma delta epsilon zeta chi
1 C -139.5 -136.7 50.9 137.6 --- --- -154.3
2 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7
3 C -139.5 -136.8 50.9 137.6 -96.5 82.0 -154.3
4 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7
5 C -139.5 -136.8 50.8 137.6 -96.5 82.0 -154.3
6 G 52.0 179.0 -173.8 94.9 -103.6 -64.8 58.7
7 C -139.5 -136.7 50.9 137.6 -96.5 81.9 -154.3
8 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7
9 C -139.5 -136.8 50.9 137.6 -96.5 82.0 -154.3
10 G --- 179.0 -173.8 94.9 -103.6 -64.8 58.7
Вывод. Самые отклоняющиеся углы:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| A-форма |
-51.70 |
174.80 |
41.70 |
79.08 |
-147.7 |
-75.0 |
-157.2 |
| B-форма |
-29.9 |
136.3 |
31.2 |
143.3 |
-140.8 |
-160.5 |
-98 |
| Z-форма |
-139.5 или 52 |
-136.7 или 179 |
-173.8 или 50.9 |
94.9 или 137.6 |
-100.4 |
-64.8 или 82 |
-154.7 или 58.7 |
Определение значения торсионных углов в заданной тРНК (1il2).
Средние значения по анализу find_pair и analyze:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| 1IL2 |
-55.4 |
56.4 |
47.8 |
87.46 |
-151.82 |
-71.27 |
-150.81 |
Strand I
base alpha beta gamma delta epsilon zeta chi
1 U --- 177.0 42.4 82.2 -145.7 -77.5 -173.0
2 C -56.3 167.0 55.1 83.8 -156.0 -76.5 -163.8
3 C -62.4 179.2 45.5 81.8 -153.6 -71.4 -159.6
4 G -62.9 174.7 50.0 79.7 -147.4 -73.0 -164.3
5 U -61.8 170.9 53.7 82.6 -147.0 -81.1 -172.2
6 G -65.5 164.4 59.9 81.9 -155.8 -76.2 -161.8
7 A -52.4 177.6 61.5 149.4 --- --- -126.3
8 c --- 133.4 29.7 79.3 -153.1 -81.2 -169.0
9 G -75.8 -176.1 51.4 84.2 -159.4 -69.5 -163.0
10 G -65.0 -175.9 46.2 81.5 -153.4 -67.0 -159.4
11 G -62.2 164.6 57.1 78.9 -155.5 -68.8 -170.3
12 G -57.3 175.9 54.6 81.8 -155.8 -65.5 -165.0
13 u -69.0 -174.2 46.4 80.5 -139.4 -68.9 -151.9
14 P -53.9 169.3 50.4 87.5 --- --- -142.0
15 C --- 162.6 -176.4 83.0 -152.6 -55.8 -159.4
16 G -71.5 -174.1 47.7 81.8 -151.6 -69.0 -158.6
17 U -54.0 167.3 52.7 85.7 -153.5 -76.5 -161.6
18 G -84.1 -176.3 52.4 82.3 -161.3 -74.7 -161.6
19 C -73.1 175.0 58.6 82.6 -159.1 -66.8 -160.4
20 C -65.0 -174.0 50.3 82.7 -155.6 -72.3 -164.0
21 A -73.1 175.9 59.1 81.5 --- --- -163.8
22 G --- 167.0 51.0 84.9 -149.9 -61.6 -178.9
23 U -70.1 179.2 58.3 84.9 -146.4 -69.1 -165.9
24 U -78.1 174.2 53.9 83.2 -160.0 -46.0 -158.0
25 P -86.8 175.3 78.2 84.1 --- --- -133.8
26 U --- -149.9 49.9 88.0 --- --- -171.6
27 A --- -166.8 45.5 78.7 -148.2 -69.7 -161.2
28 A -54.2 174.7 47.9 83.0 -156.7 -77.9 -156.3
29 u -43.8 167.2 41.1 82.5 --- --- -141.8
30 G --- -140.4 44.5 145.3 --- --- -59.2
31 C --- 167.8 118.4 84.5 -146.0 -64.3 -168.5
32 C -58.7 -175.7 36.2 80.5 -161.4 -68.8 -148.3
33 G -55.5 171.9 53.4 82.8 -145.1 -68.7 -168.6
34 U -61.0 164.6 51.6 81.8 -160.4 -73.8 -162.2
35 G -63.2 177.0 56.2 85.7 -161.8 -90.3 -162.4
36 A -46.0 159.4 60.7 140.3 --- --- -139.4
37 c --- 159.6 40.9 83.8 -151.7 -73.2 -175.4
38 G -78.7 -170.8 50.7 83.9 -160.0 -67.5 -166.2
39 G -72.2 -179.9 52.5 77.6 -153.4 -66.8 -165.5
40 G -56.7 169.3 57.1 80.6 -160.2 -68.6 -167.3
41 G -59.7 179.5 53.6 80.8 -153.7 -67.9 -165.5
42 u -65.8 179.6 51.7 82.6 -131.1 -71.0 -160.5
43 P -57.3 161.5 51.4 83.8 --- --- -150.6
44 C --- 166.0 -167.8 83.3 -135.1 -76.1 -159.3
45 G -50.0 160.7 48.5 82.9 -149.6 -63.3 -172.3
46 U -63.2 -176.3 48.6 84.1 -169.6 -73.2 -148.0
47 G -74.0 -169.5 46.8 82.9 -155.9 -75.1 -162.4
48 C -56.6 167.2 51.2 82.4 -156.9 -67.8 -163.3
49 C -67.2 179.9 51.0 82.9 -155.8 -71.5 -158.6
50 A -71.5 173.9 60.5 82.7 --- --- -167.6
51 G --- 165.2 49.7 85.4 -150.9 -64.5 -178.6
52 U -73.9 -173.4 57.8 89.4 -158.1 -63.7 -161.0
53 U -83.2 180.0 54.1 82.6 -155.4 -49.3 -152.7
54 P -82.6 168.7 84.7 82.9 --- --- -143.1
55 U --- -155.6 50.7 82.3 --- --- -160.0
56 A --- -168.7 45.6 81.8 -155.3 -64.8 -161.5
57 A -64.5 179.0 50.0 84.6 -136.7 -98.6 -151.0
58 u -60.9 142.6 60.4 85.3 --- --- -140.6
59 G --- -137.9 68.4 150.9 --- --- -57.5
Strand II
base alpha beta gamma delta epsilon zeta chi
1 A -66.6 -171.1 50.1 82.4 --- --- -163.6
2 G -63.1 164.0 61.3 80.5 -160.9 -69.1 -167.5
3 G -64.3 178.1 47.1 81.9 -150.0 -72.4 -159.4
4 C -63.3 177.0 50.9 81.8 -152.6 -72.0 -157.5
5 G -59.9 179.0 49.2 79.5 -153.7 -66.3 -158.5
6 C -64.4 164.0 53.7 79.7 -157.0 -81.5 -161.4
7 U -64.0 180.0 50.6 82.1 -138.8 -79.8 -161.9
8 G -64.5 168.1 56.4 82.9 -151.2 -71.0 -169.7
9 C -62.3 166.0 57.2 81.5 -145.4 -74.9 -164.4
10 C -62.2 175.9 50.5 80.0 -148.0 -72.5 -163.8
11 C -65.0 -178.4 44.7 81.3 -159.9 -68.5 -153.0
12 C --- -165.7 42.9 81.8 -150.1 -75.0 -168.0
13 A --- -127.8 58.5 146.6 --- --- -75.6
14 G --- -179.8 -55.8 140.9 --- --- -113.5
15 P -63.0 176.1 44.9 85.5 --- --- -151.5
16 C -45.6 157.9 41.5 78.6 -148.4 -64.2 -168.2
17 G 150.7 -177.7 -174.5 86.5 -123.4 -82.0 177.1
18 C -58.1 -176.5 39.5 83.5 -176.1 -61.5 -149.0
19 G -59.4 -174.6 41.0 82.5 -150.7 -71.0 -162.0
20 G -72.5 -164.8 47.2 81.6 -154.6 -71.8 -168.9
21 G -69.4 175.8 54.3 79.8 -153.6 -63.6 -160.9
22 U -64.4 166.0 54.8 85.0 -157.6 -57.5 -164.9
23 A -59.4 162.7 50.8 82.0 -145.6 -71.3 -154.7
24 A -51.2 163.2 58.5 80.8 -147.4 -81.8 -158.5
25 G --- 155.3 46.3 80.3 -143.1 -79.7 -167.5
26 A --- 165.1 56.5 84.6 --- --- -128.7
27 A --- -167.0 53.4 86.0 --- --- -151.0
28 U --- 126.9 167.5 142.5 --- --- -126.9
29 U --- -150.3 65.6 87.8 --- --- -173.5
30 C --- 159.5 55.6 82.4 --- --- -179.0
31 G -58.9 -170.6 46.5 83.0 --- --- -144.6
32 G -62.0 176.3 49.5 82.8 -157.1 -63.1 -166.5
33 C -59.0 160.3 53.4 82.4 -149.2 -76.8 -168.3
34 G 160.2 -178.7 176.6 82.8 -132.8 -80.5 -179.7
35 C -51.7 163.1 42.4 81.3 -174.7 -67.8 -156.2
36 U -40.6 161.4 42.0 83.3 -143.2 -78.7 -161.0
37 G -52.3 164.8 55.0 80.2 -147.3 -81.7 -168.1
38 C -51.3 171.9 45.1 77.2 -146.6 -73.8 -160.8
39 C -70.2 -173.3 51.4 81.1 -149.0 -75.2 -162.3
40 C -65.4 172.9 52.4 80.8 -154.9 -72.7 -169.0
41 C --- -176.9 55.7 82.0 -145.0 -78.3 -167.6
42 A --- -151.9 51.9 145.9 --- --- -86.0
43 G --- 148.2 -166.8 81.3 --- --- -162.9
44 P -56.3 175.6 43.5 87.8 --- --- -154.0
45 C -46.1 175.3 36.1 80.1 -149.3 -64.4 -159.4
46 G 152.9 -168.6 178.0 83.9 -119.7 -75.4 167.6
47 C -53.4 178.7 40.2 83.9 -173.8 -77.0 -147.5
48 G -71.8 -169.4 50.6 82.8 -149.0 -71.3 -156.6
49 G -65.4 -168.3 45.1 81.2 -154.0 -74.0 -172.9
50 G -77.0 -178.1 55.7 82.4 -152.3 -63.7 -166.9
51 U -69.1 161.3 60.1 84.7 -156.6 -55.8 -161.9
52 A -68.4 173.7 53.1 84.9 -149.0 -73.5 -158.4
53 A -52.5 173.2 48.4 82.2 -155.4 -74.6 -159.0
54 G --- 145.5 48.0 82.2 -148.4 -79.0 -168.1
55 A --- 162.0 60.6 85.8 --- --- -128.2
56 A --- 133.3 31.4 83.6 --- --- -152.6
57 U --- 125.9 175.0 143.1 --- --- -126.7
58 U --- -149.6 59.9 88.2 --- --- -165.1
59 C --- 164.0 53.8 83.6 --- --- -171.4
Вывод. Данная структура больше всего похожа на А-форму.
Определение торсионных углов в заданной структре ДНК (1odh).
Средние значения по анализу find_pair и analyze:
|
α (P - O5') |
β (O5' - C5') |
γ (C5' - C4') |
δ (C4' - C3') |
ε (C3' - O3') |
ξ (O3' - P) |
χ (C1' - N) |
| 1ODH |
-25.99 |
30.19 |
26.57 |
141.93 |
-39.92 |
-100.23 |
-109.84 |
Strand I
base alpha beta gamma delta epsilon zeta chi
1 C --- --- -60.1 147.1 162.3 -85.8 -106.3
2 G 158.4 -158.8 169.0 139.5 -155.5 -144.4 -122.7
3 A -40.4 165.2 37.1 147.1 -169.0 -110.8 -105.0
4 T 35.9 179.0 -61.3 151.8 -171.0 -127.1 -96.7
5 G 29.4 167.9 -46.6 155.1 165.4 -90.4 -103.1
6 C -50.7 -164.5 36.4 136.5 -156.4 -106.8 -110.5
7 G -60.0 163.7 40.5 148.1 -112.2 166.2 -96.3
8 G -59.5 142.0 37.5 135.6 -168.9 -87.9 -128.8
9 G -68.7 179.9 38.0 132.4 171.7 -117.0 -113.9
10 T -34.6 -171.4 35.0 141.1 -168.0 -108.4 -117.1
11 G -54.8 175.0 38.6 136.7 -177.4 -124.5 -112.3
12 C 43.1 174.1 -51.2 151.9 167.1 -82.1 -109.1
13 A 170.0 147.3 172.8 149.0 --- --- -144.1
Strand II
base alpha beta gamma delta epsilon zeta chi
1 G -61.4 151.9 42.0 143.1 --- --- -99.6
2 C -34.4 156.8 31.1 147.0 -132.6 -162.6 -96.8
3 T -39.5 -179.1 43.3 148.0 -162.1 -117.3 -107.2
4 A -56.6 -167.3 37.9 139.0 173.7 -118.1 -112.2
5 C -58.5 -162.0 40.0 139.7 175.4 -92.6 -112.5
6 G -26.7 150.9 44.9 140.2 175.2 -105.8 -117.1
7 C -85.0 -142.0 45.0 142.4 -179.7 -137.0 -106.2
8 C -40.9 156.6 28.1 133.0 164.0 -81.1 -120.1
9 C -38.5 168.2 29.6 132.9 -148.8 -153.5 -105.3
10 A -47.6 165.4 39.6 137.0 -169.1 -105.0 -112.9
11 C -66.1 -162.8 38.2 137.2 -165.8 -124.4 -106.6
12 G 24.0 -172.7 -66.2 150.3 165.8 -74.4 -104.0
13 T --- --- 172.6 150.1 -154.2 -126.6 -98.5
Вывод. Самое "деформированное" основание, на первый взгляд, второй аденин. Выделен цветом.
Определение номеров нуклеотидов, образующих стебли (stems) в тРНК.
| Акцепторный стебель |
T-стебель |
Антикодоновый стебель |
D-стебель |
| 901 - 972 |
949 - 965 |
938 - 932 |
910 - 925 |
| 907 - 966 |
953 - 961 |
944 - 926 |
913 - 922 |
Водородные связи из .out:
(----- for WC bp, + for isolated bp, x for helix change)
Strand I Strand II Helix
1 (0.003) C:.901_:[..U]U-----A[..A]:.972_:C (0.005) |
2 (0.004) C:.902_:[..C]C-----G[..G]:.971_:C (0.009) |
3 (0.004) C:.903_:[..C]C-----G[..G]:.970_:C (0.011) |
4 (0.012) C:.904_:[..G]G-----C[..C]:.969_:C (0.004) |
5 (0.008) C:.905_:[..U]U-*---G[..G]:.968_:C (0.010) |
6 (0.006) C:.906_:[..G]G-----C[..C]:.967_:C (0.008) |
7 (0.006) C:.907_:[..A]Ax----U[..U]:.966_:C (0.004) |
8 (0.017) C:.949_:[5MC]c-----G[..G]:.965_:C (0.008) |
9 (0.005) C:.950_:[..G]G-----C[..C]:.964_:C (0.005) |
10 (0.006) C:.951_:[..G]G-----C[..C]:.963_:C (0.006) |
11 (0.012) C:.952_:[..G]G-----C[..C]:.962_:C (0.005) |
12 (0.009) C:.953_:[..G]G----xC[..C]:.961_:C (0.008) |
13 (0.012) C:.954_:[5MU]u-**-xA[..A]:.958_:C (0.007) |
14 (0.043) C:.955_:[PSU]Px**+xG[..G]:.917_:C (0.005) x
15 (0.005) C:.938_:[..C]C-*---P[PSU]:.932_:C (0.044) |
16 (0.006) C:.939_:[..G]G-----C[..C]:.931_:C (0.006) |
17 (0.003) C:.940_:[..U]U-*---G[..G]:.930_:C (0.009) |
18 (0.010) C:.941_:[..G]G-----C[..C]:.929_:C (0.004) |
19 (0.010) C:.942_:[..C]C-----G[..G]:.928_:C (0.004) |
20 (0.003) C:.943_:[..C]C-----G[..G]:.927_:C (0.005) |
21 (0.005) C:.944_:[..A]Ax*---G[..G]:.926_:C (0.005) |
22 (0.009) C:.910_:[..G]G-*---U[..U]:.925_:C (0.004) |
23 (0.003) C:.911_:[..U]U-----A[..A]:.924_:C (0.009) |
24 (0.007) C:.912_:[..U]U-----A[..A]:.923_:C (0.007) |
25 (0.044) C:.913_:[PSU]Px*--xG[..G]:.922_:C (0.005) |
26 (0.003) C:.908_:[..U]Ux**-xA[..A]:.946_:C (0.005) |
27 (0.003) C:.914_:[..A]A-*--xA[..A]:.921_:C (0.008) |
28 (0.004) C:.915_:[..A]A-**+xU[..U]:.948_:C (0.003) |
29 (0.115) C:.916_:[H2U]ux**+xU[..U]:.959_:C (0.006) x
30 (0.021) C:.918_:[..G]Gx---xC[..C]:.956_:C (0.007) +
Площади перекрывания пар:
step i1-i2 i1-j2 j1-i2 j1-j2 sum
1 UC/GA 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 2.52( 1.02) 2.52( 1.02)
2 CC/GG 0.00( 0.00) 0.00( 0.00) 0.27( 0.00) 4.35( 3.12) 4.62( 3.12)
3 CG/CG 0.07( 0.00) 0.00( 0.00) 4.34( 1.58) 0.00( 0.00) 4.41( 1.58)
4 GU/GC 6.93( 4.29) 0.00( 0.00) 0.00( 0.00) 6.95( 3.82) 13.88( 8.11)
5 UG/CG 0.04( 0.00) 0.00( 0.00) 4.43( 1.56) 0.00( 0.00) 4.47( 1.56)
6 GA/UC 3.95( 2.44) 0.00( 0.00) 0.00( 0.00) 1.52( 0.32) 5.47( 2.76)
7 Ac/GU 3.57( 0.85) 0.00( 0.00) 0.00( 0.00) 6.18( 3.23) 9.75( 4.09)
8 cG/CG 0.00( 0.00) 0.00( 0.00) 3.86( 1.41) 0.39( 0.01) 4.24( 1.42)
9 GG/CC 2.27( 0.85) 0.00( 0.00) 0.26( 0.00) 1.27( 0.37) 3.80( 1.22)
10 GG/CC 5.13( 3.77) 0.00( 0.00) 0.11( 0.00) 0.38( 0.00) 5.62( 3.77)
11 GG/CC 3.90( 2.52) 0.00( 0.00) 0.65( 0.00) 0.00( 0.00) 4.55( 2.52)
12 Gu/AC 8.16( 3.29) 0.00( 0.00) 0.00( 0.00) 5.49( 2.41) 13.66( 5.70)
13 uP/GA 5.69( 1.65) 0.00( 0.00) 0.00( 0.00) 6.67( 3.61) 12.36( 5.26)
14 PC/PG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
15 CG/CP 0.64( 0.30) 0.00( 0.00) 0.00( 0.00) 3.57( 1.51) 4.21( 1.81)
16 GU/GC 7.38( 3.96) 0.00( 0.00) 0.00( 0.00) 7.11( 4.09) 14.49( 8.06)
17 UG/CG 0.00( 0.00) 0.00( 0.00) 6.00( 3.08) 0.00( 0.00) 6.00( 3.08)
18 GC/GC 5.84( 2.75) 0.00( 0.00) 0.00( 0.00) 5.42( 2.57) 11.26( 5.33)
19 CC/GG 0.05( 0.00) 0.00( 0.00) 0.55( 0.00) 1.95( 0.55) 2.55( 0.55)
20 CA/GG 0.38( 0.00) 0.00( 0.00) 1.45( 0.51) 1.18( 0.18) 3.01( 0.69)
21 AG/UG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.38( 0.14) 0.38( 0.14)
22 GU/AU 6.73( 3.84) 0.00( 0.00) 0.00( 0.00) 5.58( 4.52) 12.31( 8.36)
23 UU/AA 1.31( 0.02) 0.00( 0.00) 0.00( 0.00) 2.24( 1.93) 3.55( 1.96)
24 UP/GA 1.76( 0.24) 0.00( 0.00) 0.00( 0.00) 3.67( 2.38) 5.43( 2.63)
25 PU/AG 2.99( 0.73) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 2.99( 0.73)
26 UA/AA 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 3.31( 2.31) 3.31( 2.31)
27 AA/UA 5.89( 3.47) 0.00( 0.00) 0.00( 0.00) 4.87( 2.39) 10.76( 5.86)
28 Au/UU 5.28( 3.84) 0.00( 0.00) 0.00( 0.00) 4.70( 0.64) 9.98( 4.48)
29 uG/CU 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
30 GC/GC 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
31 CC/GG 0.00( 0.00) 0.00( 0.00) 0.27( 0.00) 3.38( 2.02) 3.65( 2.02)
32 CG/CG 0.00( 0.00) 0.00( 0.00) 4.91( 2.11) 0.00( 0.00) 4.91( 2.11)
33 GU/GC 7.25( 4.31) 0.00( 0.00) 0.00( 0.00) 6.58( 3.48) 13.84( 7.79)
34 UG/CG 0.00( 0.00) 0.00( 0.00) 5.98( 3.15) 0.00( 0.00) 5.98( 3.15)
35 GA/UC 3.17( 1.29) 0.00( 0.00) 0.00( 0.00) 1.50( 0.18) 4.66( 1.47)
36 Ac/GU 5.06( 1.54) 0.00( 0.00) 0.00( 0.00) 7.01( 4.34) 12.08( 5.88)
37 cG/CG 0.00( 0.00) 0.00( 0.00) 4.39( 1.52) 0.14( 0.00) 4.53( 1.53)
38 GG/CC 3.13( 1.64) 0.00( 0.00) 0.44( 0.00) 0.48( 0.07) 4.05( 1.71)
39 GG/CC 4.49( 2.86) 0.00( 0.00) 0.53( 0.00) 0.00( 0.00) 5.02( 2.86)
40 GG/CC 3.22( 1.74) 0.00( 0.00) 0.82( 0.00) 0.00( 0.00) 4.04( 1.74)
41 Gu/AC 8.53( 3.99) 0.00( 0.00) 0.00( 0.00) 5.60( 2.62) 14.13( 6.61)
42 uP/GA 6.11( 1.84) 0.00( 0.00) 0.00( 0.00) 7.23( 4.69) 13.34( 6.52)
43 PC/PG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
44 CG/CP 1.12( 0.64) 0.00( 0.00) 0.00( 0.00) 3.64( 1.58) 4.76( 2.22)
45 GU/GC 6.81( 3.55) 0.00( 0.00) 0.00( 0.00) 5.82( 2.76) 12.63( 6.31)
46 UG/CG 0.00( 0.00) 0.00( 0.00) 5.72( 2.85) 0.00( 0.00) 5.72( 2.85)
47 GC/GC 6.67( 3.64) 0.00( 0.00) 0.00( 0.00) 7.00( 3.98) 13.67( 7.62)
48 CC/GG 0.83( 0.04) 0.00( 0.00) 0.12( 0.00) 2.10( 0.67) 3.04( 0.71)
49 CA/GG 0.42( 0.00) 0.00( 0.00) 1.33( 0.60) 1.21( 0.17) 2.95( 0.77)
50 AG/UG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.07( 0.01) 0.07( 0.01)
51 GU/AU 6.65( 3.90) 0.00( 0.00) 0.00( 0.00) 5.12( 4.12) 11.77( 8.02)
52 UU/AA 1.05( 0.01) 0.00( 0.00) 0.00( 0.00) 2.84( 2.43) 3.88( 2.44)
53 UP/GA 1.86( 0.20) 0.00( 0.00) 0.00( 0.00) 2.99( 1.58) 4.85( 1.78)
54 PU/AG 3.09( 0.55) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 3.09( 0.55)
55 UA/AA 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 4.02( 2.90) 4.02( 2.90)
56 AA/UA 6.10( 3.99) 0.00( 0.00) 0.00( 0.00) 6.46( 3.76) 12.56( 7.75)
57 Au/UU 4.65( 2.82) 0.00( 0.00) 0.00( 0.00) 2.76( 0.05) 7.41( 2.86)
58 uG/CU 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
Пары с наибольшей площадью перекрывания (выделены красным цветом в списке выше):
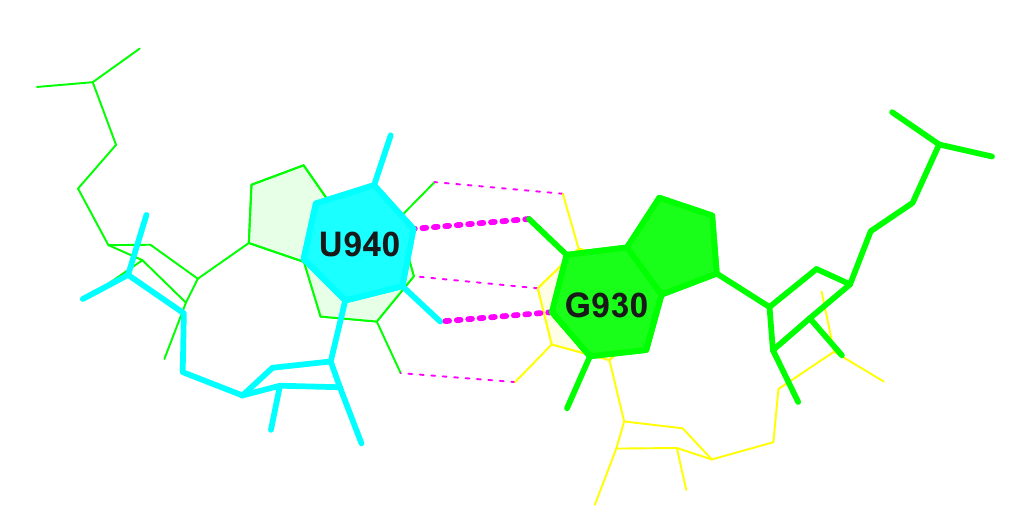
Соответствующая строка из файла stacking.pdb:
ATOM 16 N3 U C 901 0.291 3.315 -1.415 1.00 60.88 N
Изображение для пары с малой площадью перекрытия (выбранная обозначена синим в списке):
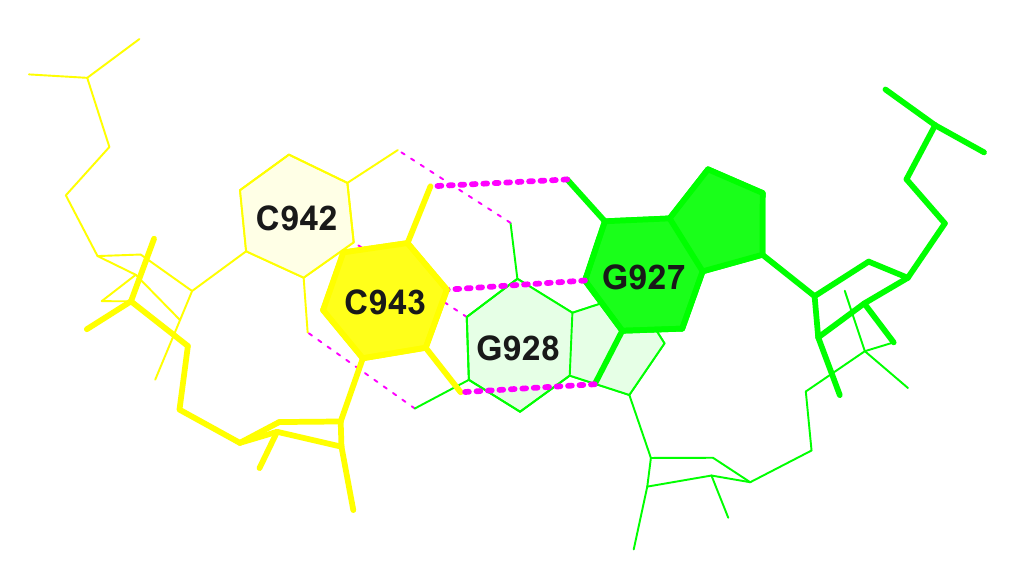
Соответствующая строка из файла stacking.pdb:
ATOM 19 C5 U C 901 2.120 4.814 -1.385 1.00 55.97 C
Взаимную ориентацию оснований также можно посмотреть в RasMol, вырезав основания, соседние к указанным в соответствующих строках stacking.pdb

