Protein description
Chitinases belong to the class of glycosyl hydrolases that degrade chitin, an abundant natural polysaccharide, by cleaving the β-(1,4)
linkages of its N-acetylglucosamine chain (Gooday, 1990). Our protein (PDB ID: 4WKH) is the catalytic domain of one of such chitinases,
chitotriosidase-1 (CHIT1), which belongs to the highly conserved glycosyl hydrolase family 18 (GH18). It is one of the two active
chitinases that have been identified in humans and have been reported to be involved in the innate immune response as well as being
a biomarker of Gaucher disease (Hollak et al.,1994) (this lysosomal enzyme is very useful for monitoring Gaucher disease activity in response
to treatment, and may reflect the severity of the disease). Gaucher disease is a genetic disorder in which glucocerebroside accumulates
in cells and certain organs. The disorder is characterized by bruising, fatigue, anemia,low blood platelet count and enlargement of the liver and spleen,
and is caused by a hereditary deficiency of the enzyme glucocerebrosidase (also known as glucosylceramidase), which acts on glucocerebroside.
Here is the hydrolysis mechanism of CHIT1 proposed by Fadel et al., 2015:
 You can look at the catalytic center via the Jmol applet ("Interactions with Ligand" button) (several configurations are superimposed there,
which gives rise to some amino acids being "double-headed"). The ligand is a monomer of chitin.
You can look at the catalytic center via the Jmol applet ("Interactions with Ligand" button) (several configurations are superimposed there,
which gives rise to some amino acids being "double-headed"). The ligand is a monomer of chitin.
Contacts identification process
For hydrogen bonding identification secondary structure was first analyzed and then we picked sites where hydrogen bonds were not parts of any kind of secondary
helices or β-sheets. In order to spot salt bridges and hydrophobic contacts we looked for charged amino acids and hydrophobic ones, respectively.
Disulfide bridges were identified by spotting all cysteines. Interactions with the ligand were found by gradually reducing the area selected around the ligand.
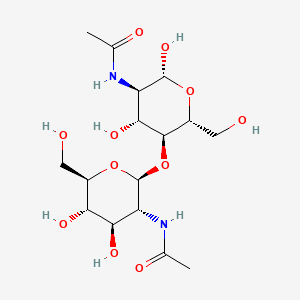
Ligand:
Here interactions with the protein are shown:

Measurements

For hydrogen bonding identification secondary structure was first analyzed and then we picked sites where hydrogen bonds were not parts of any kind of secondary
helices or β-sheets. In order to spot salt bridges and hydrophobic contacts we looked for charged amino acids and hydrophobic ones, respectively.
Disulfide bridges were identified by spotting all cysteines. Interactions with the ligand were found by gradually reducing the area selected around the ligand.
Ligand:
Here interactions with the protein are shown:


Measurements
| Atom labels | Distance (Å) | Angle (°) |
| Hydrogen bonds | ||
| N(296:A LYS)-O(292:A GLY) | 3.05 | 128.0 |
| N(228:A LEU)-O(320:A GLN) | 2.98 | 143.9 |
| N(10:A SER)-O(6:A GLY) | 3.16 | 130.5 |
| Salt bridges | ||
| Atom labels | Distance (Å) | |
| OD2(205:A ASP)-NH2(177:A ARG) | 3.79 | |
| OD1(205:A ASP)-NH1(177:A ARG) | 3.39 | |
| OD1(205:A ASP)-NH2(177:A ARG) | 2.74 | |
| OD2(205:A ASP)-NH1(177:A ARG) | 2.90 | |
| OE1(169:A GLU)-NH2(128:A ARG) | 2.87 | |
| OE2(169:A GLU)-NE(128:A ARG) | 2.85 | |
| OD2(133:A ASP)-NZ(84:A LYS) | 2.85 | |
| OD1(133:A ASP)-NZ(84:A LYS) | 3.41 | |
| Disulfide bridges | ||
| SG(51:A CYS)-SG(26:A CYS) | 2.85 | |
| SG(370:A CYS)-SG(307:A CYS) | 3.41 | |
Ligand Info
| IUPAC Name | N-[(2R,3R,4R,5S,6R)-5-[(2S,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-2,4-dihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide | |
| Molecular formula | C16H28N2O11 | |
| Molar weight | 424.403 g/mol | |
| Page at PubChem | ||
Hydrophobic core
The analysis of the hydrophobic core was carried out. CluD hydrophobic cluster analysis service showed presence of 6 hydrophobic cores,
but all of them, except the first one, included less than 10 atoms, so they were not taken into account.The results of
the work can be seen in the applet window and in the images below.Use "Core" and "Resume(Core)" buttons to navigate through the applet.
The density of atoms in the core.

From the figure above it is possible to infer that the larger the distance from the residue the more atoms fall within this distance. However, the increase
is not uniform: at distances 3-4 and 4-5 angstroms there are considerably more atoms than it should be with such
course of the curve. With this fact taken into account it is possible to deduce that distribution of hydrophobic amino acids's atoms in the core
is not random.
The minimum distance
It should be pointed out that the residue surface can be seen even when all the atoms (represented with their van der Waals radii) within the distance of 7 angstroms clutter the model.
Because of the lack of specifity of the "atoms should be barely seen" criterion, it was decided to keep the distance of 6 angstroms. Atoms at the
distance of 6 angstroms from the residue cover its surface in such a way that it can be barely seen.
Typical distance
From the diagram given above it can be seen that with the distance increasing from 3 to 5 angstroms a steep increase in number of atoms occurs. The largest
residue surface is covered on the distance from 3 to 5 angstroms, which can be seen in the applet. At the same time covalently bound disappear at the
distance from 2 to 3 angstroms, which also can be seen in the applet. That is why it possible to make a conclusion that a typical distance between
unbound atoms in the protein is smaller than 5 angstroms, beeing about 3-4 angstroms.
Water molecule
Taking into account all the information above, making a conclusion about the possibility of existence of a water molecule between neghbouring atoms of the protein's
hydrophobic core becomes trivial. It is clear that the water molecule will be trapped between two atoms, if the sum of the radii of these two atoms, with the diameter of the
water molecule added is less than the characteristic distance. But even with the "optimistic" calculations of the sum above it is about 5.6 angstroms, which is considerably
bigger that it should be to make water molecule stuck between the atoms of the protein.
Author contributions
ST together with SK found and analyzed protein contacts, composed
Jmol scripts responsible for visualising them. ST created the visual model of the ligand, its interactions with the protein,
analyzed the protein's biological role and made the description in English and Tatar.
SK translated the protein description to Russian, translated the hydrophobic core part
to English with assistance of ST.
RG analyzed the parameters of the hydrophobic core and wrote their description in Russian, constructed CSS/HTML code of the webpage.
References
[1]Gooday, G. (1990). Adv. Microb. Ecol. 11, 387-430.
[2]Hollak, C. E., van Weely, S., van Oers, M. H. & Aerts, J. M. (1994). J. Clin. Invest. 93, 1288-1292.
[3]Fadel, F., Zhao, Y., Cachau, R., Cousido-Siah, A., Ruiz, F.X., Harlos, K., Howard, E., Mitschler, A., Podjarny, A. (2015). Acta Cryst. 71, 1455-1470.

The minimum distance
It should be pointed out that the residue surface can be seen even when all the atoms (represented with their van der Waals radii) within the distance of 7 angstroms clutter the model.
Because of the lack of specifity of the "atoms should be barely seen" criterion, it was decided to keep the distance of 6 angstroms. Atoms at the
distance of 6 angstroms from the residue cover its surface in such a way that it can be barely seen.
Typical distance
From the diagram given above it can be seen that with the distance increasing from 3 to 5 angstroms a steep increase in number of atoms occurs. The largest
residue surface is covered on the distance from 3 to 5 angstroms, which can be seen in the applet. At the same time covalently bound disappear at the
distance from 2 to 3 angstroms, which also can be seen in the applet. That is why it possible to make a conclusion that a typical distance between
unbound atoms in the protein is smaller than 5 angstroms, beeing about 3-4 angstroms.
Water molecule
Taking into account all the information above, making a conclusion about the possibility of existence of a water molecule between neghbouring atoms of the protein's
hydrophobic core becomes trivial. It is clear that the water molecule will be trapped between two atoms, if the sum of the radii of these two atoms, with the diameter of the
water molecule added is less than the characteristic distance. But even with the "optimistic" calculations of the sum above it is about 5.6 angstroms, which is considerably
bigger that it should be to make water molecule stuck between the atoms of the protein.
Author contributions
ST together with SK found and analyzed protein contacts, composed
Jmol scripts responsible for visualising them. ST created the visual model of the ligand, its interactions with the protein,
analyzed the protein's biological role and made the description in English and Tatar.
SK translated the protein description to Russian, translated the hydrophobic core part
to English with assistance of ST.
RG analyzed the parameters of the hydrophobic core and wrote their description in Russian, constructed CSS/HTML code of the webpage.
References
[1]Gooday, G. (1990). Adv. Microb. Ecol. 11, 387-430.
[2]Hollak, C. E., van Weely, S., van Oers, M. H. & Aerts, J. M. (1994). J. Clin. Invest. 93, 1288-1292.
[3]Fadel, F., Zhao, Y., Cachau, R., Cousido-Siah, A., Ruiz, F.X., Harlos, K., Howard, E., Mitschler, A., Podjarny, A. (2015). Acta Cryst. 71, 1455-1470.
SK translated the protein description to Russian, translated the hydrophobic core part to English with assistance of ST.
RG analyzed the parameters of the hydrophobic core and wrote their description in Russian, constructed CSS/HTML code of the webpage.
[3]Fadel, F., Zhao, Y., Cachau, R., Cousido-Siah, A., Ruiz, F.X., Harlos, K., Howard, E., Mitschler, A., Podjarny, A. (2015). Acta Cryst. 71, 1455-1470.