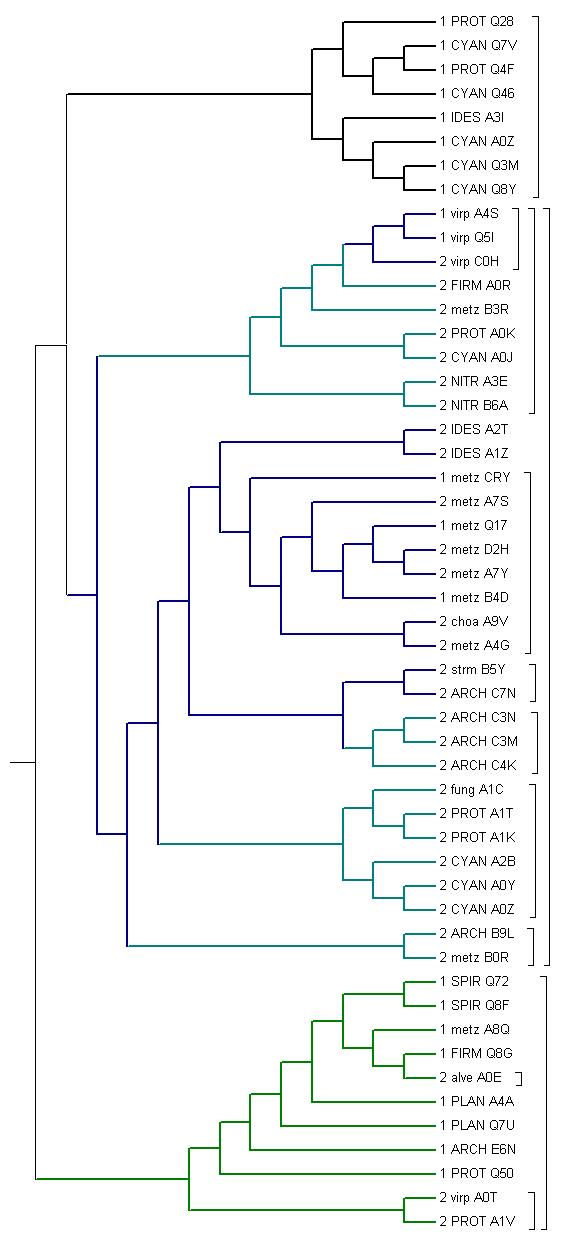
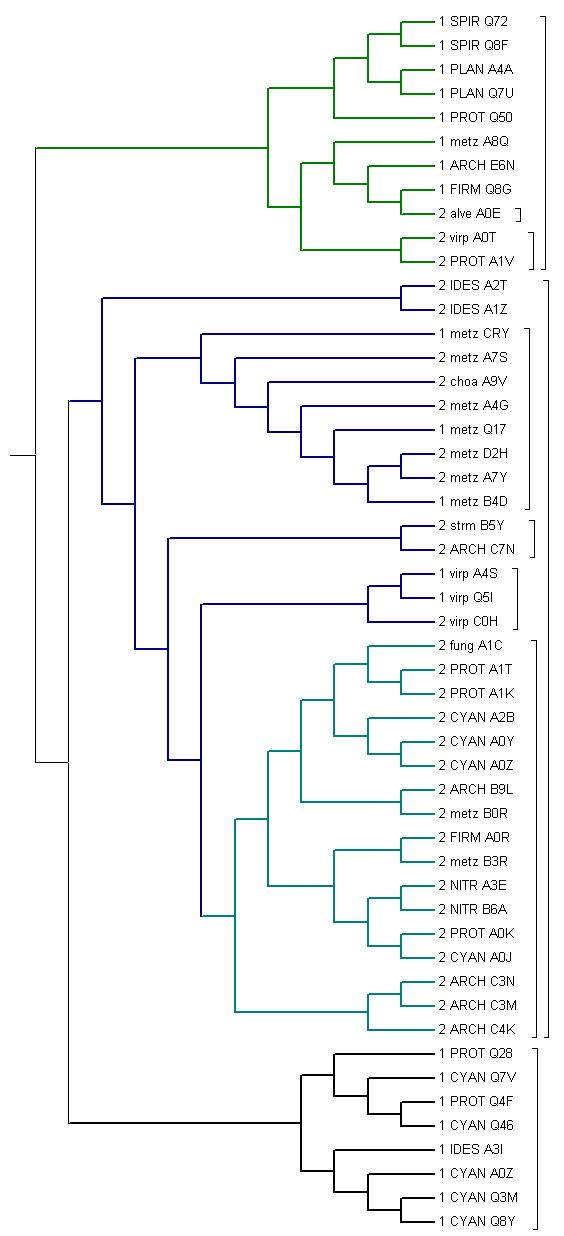
мелким шрифтом обозначены эукариоты, крупным-бактерии и археи 1-однодоменная архитектура 2-двухдоменная архитектура ARCH Archaea virp Viridiplantae metz Metazoa alve Alveolata fung FUNGI strm Stramenophyla choa Choanoflagellida PROT Proteobacteria CYAN Cyanobacteria IDES Bacteroidetes FIRM Firmicutes NITR Nitrospirae SPIR Spirochaetes PLAN Planctomycetes
+-----------------------1 metz Q17
|
+--67.0-| +-------2 metz D2H
| | +--100.0-|
| +--88.8-| +-------2 metz A7Y
+--38.0-| |
| | +---------------1 metz B4D
| |
+--50.7-| | +-------2 choa A9V
| | +------------------58.0-|
| | +-------2 metz A4G
+--72.0-| |
| | +---------------------------------------2 metz A7S
| |
+--56.3-| +-----------------------------------------------1 metz CRY
| |
| | +-------2 IDES A2T
| +------------------------------------------100.0-|
| +-------2 IDES A1Z
|
| +-------2 NITR A3E
| +--------------------------100.0-|
| | +-------2 NITR B6A
| |
+--13.2-| | +-------2 FIRM A0R
| | +--14.0-| +----------34.7-|
| | | | | +-------2 metz B3R
| | | | +--39.0-|
| | | | | | +-------1 virp A4S
| | | | | | +--100.0-|
| | +--21.0-| +--24.5-| +--43.5-| +-------1 virp Q5I
| | | | | |
| | | | | +---------------2 virp C0H
| | | | |
| +--17.3-| | +-------------------------------2 PROT A0K
+--43.6-| | |
| | | +-----------------------------------------------2 CYAN A0J
| | |
| | | +-------2 PROT A1K
| | +------------------------------------------98.0-|
| | +-------2 PROT A1T
| |
| | +-------2 CYAN A0Z
| | +--100.0-|
| | +--99.0-| +-------2 CYAN A0Y
+--60.0-| | | |
| | +------------------------------------------36.3-| +---------------2 CYAN A2B
| | |
| | | +---------------2 fung A1C
| | +--31.2-|
| | | +-------2 metz B0R
| | +--72.0-|
+--78.7-| | +-------2 ARCH B9L
| | |
| | | +-------2 ARCH C3N
| | | +--94.5-|
| | +----------------------------------------------------------100.0-| +-------2 ARCH C3M
+--99.0-| | |
| | | +---------------2 ARCH C4K
| | |
| | +---------------------------------------------------------------------------------------2 ARCH C7N
| |
| +-----------------------------------------------------------------------------------------------2 strm B5Y
|
+--100.0-| +-------1 CYAN Q3M
| | +--94.8-|
| | +--100.0-| +-------1 CYAN Q8Y
| | | |
| | +--57.5-| +---------------1 CYAN A0Z
| | | |
| | | +-----------------------1 IDES A3I
| | |
+--55.2-| +------------------------------------------------------------------98.0-| +-------1 PROT Q4F
| | | +--82.8-|
| | | +--89.0-| +-------1 CYAN Q7V
| | | | |
| | +--98.0-| +---------------1 CYAN Q46
+--33.1-| | |
| | | +-----------------------1 PROT Q28
| | |
| | +---------------------------------------------------------------------------------------------------------------1 ARCH E6N
| |
+--100.0-| +-----------------------------------------------------------------------------------------------------------------------2 alve A0E
| |
| | +---------------1 PROT Q50
| | +--29.6-|
| | | | +-------1 FIRM Q8G
| | | +--85.7-|
| +--------------------------------------------------------------------------------------------------21.3-| +-------1 metz A8Q
+-------| |
| | | +-------2 virp A0T
| | +----------81.0-|
| | +-------2 PROT A1V
| |
| | +-------1 SPIR Q8F
| | +--100.0-|
| +------------------------------------------------------------------------------------------------------------------70.0-| +-------1 SPIR Q72
| |
| +---------------1 PLAN A4A
|
+-----------------------------------------------------------------------------------------------------------------------------------------------1 PLAN Q7U
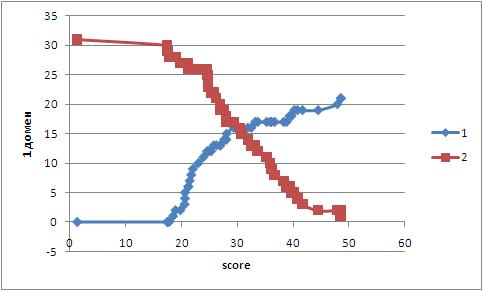
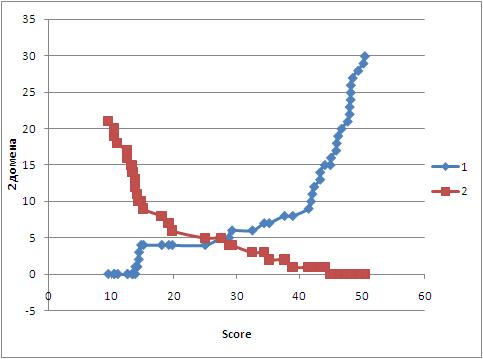
Оба профиля были исследованы на количество ошибок первого (ложноположительные) и второго (ложноотрицательные) родов.
 |  |