Applet controls:
Ligands:
Contacts:
Introduction
Tubulin is a protein that makes the basic part of the microtubule and consists of 2 molecules – α and β-tubulin (A chain [UniProt ID B6A7R0] and B chain [UniProt ID P02554]). Protein Data Base ID is 5SYC. It exists in a form of dimer within cells. We have chosen this structural image because it has the highest resolution available (0.35nm). This protein was taken from Sus scrofa's proteome.
Considering that we have to work with a complex of molecules which consists of the basic element of microtubule, we've got to describe both structure and functionality of microtubules. Other tubulin types will not be taken into account because they might be significantly different from α- and β-tubulin molecules.
Microtubules are intracellular protein structures that are comprised by cytoskeleton. Their external diameter equals about 25 nm, such size corresponds to the helical structure each coil of which comprises 13 subunits arranged. Cells are able to form microtubules more that 10 mm long. These tubules play a significant role in many transportative and regulative processes. Such an active use of microtubules by cells can be explained by some of peculiar features of their structure. The most important one is that microtubules consist of identical monomers that are able to polimerize rapidly in presence of trace amounts of GTP and form the potentially infinite structure. That provides rapid growth.[3] The second feature is their dynamic instability. Microtubule is a quite unstable aggregation. If the GTP molecule that stabilizes the tubule gets hydrolyzed, the complex will collapse quite quickly. These features help cells to assemble and degrade microtubules in a relatively short periods of time; that is useful, e.g. during cell division.[3] Cells are able to stop the process by capping microtubules and disrupting their self-disassemblement. Finally, microtubules turned out to be a perfect substrate forconnecting with various transporting proteins, which helps to implement complicated transportation.[4]
The protein has 4 different ligands:
Peroluside A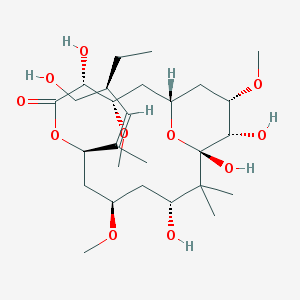 IUPAC name: (1S,3S,4R,7R,9R,11R,13S,14S,15S)-4,11,13,14-tetrahydroxy-7-[(2Z,4S)-4-(hydroxymethyl)hex-2-en-2-yl]-3,9,15-trimethoxy-12,12-dimethyl-6,17-dioxabicyclo[11.3.1]heptadecan-5-one Molecular formula: C27H48O11 Molar mass: 548.67 g/mol PubChem |
Guanosine 5’-triphosphate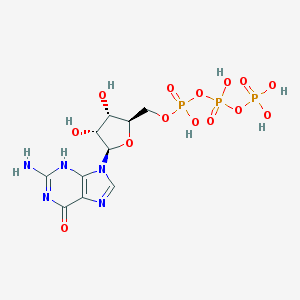 IUPAC name: [[(2R,3S,4R,5R)-5-(2-amino-6-oxo-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl] phosphono hydrogen phosphate Molecular formula: С10H16N5O14P3 Molar mass: 523.18 g/mol PubChem |
Guanosine 5’-diphosphate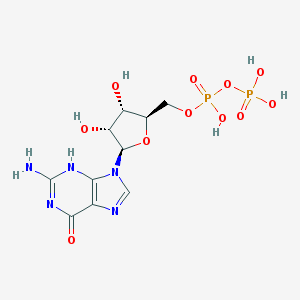 IUPAC name: [(2R,3S,4R,5R)-5-(2-amino-6-oxo-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl phosphono hydrogen phosphate Molecular formula: C10H15N5O11P2 Molar mass: 443.2 g/mol PubChem |
Magnesium (II)IUPAC name: Magnesium(2+)Molecular formula: Mg2+ Molar mass: 24.305 g/mol PubChem |
Contacts information:
One of the most important ligand-protein contacts is the contact between A chain and Mg2+. Its structure is quite obvious but not its function. Magnesium is coordinated with Glu-71 via the ionic bond. It still has one vacant valence that is used to stabilize the unstable pyrophosphate bond next to it.
We should also state that (the 3D-alignment showed no significant differences) both tubulin chains in this part (α and β) can be considered identical.
Because of a significant functional role of tubulin there are only three types of intermolecular bonds: hydrogen bonds, hydrophobic interactions, and charged interactions. Therefore, covalent intermolecular bonds are not common among tubulin protein family.
It is important to note that JMol was able to find only one intermolecular hydrogen bond; it could be a confirmation of the fact that this type of bonds play the least important role in formation of the complex. You can take a look at this connection in the applet above.
Nevertheless, a role of other types of binding is quite interesting. The most important ones could be hydrophobic interactions. As it is shown in the applet, hydrophobic core is relatively large and has several exposures. Two parts of hydrophobic cores do interact with each other.
Yet another type of contacts between tubulin units are ionic bonds. These are situated on the connecting edges of the molecules (despite being secondary), because the main function is performed by hydrophobic and ligand interactions. The most important role though is played by them on the other sides of the protein, where the protophilament chains form a microtubule. As you can see, the secondary sides are covered in charged amino acids, which are required to form ionic bonds. This assumtion gets confirmed in other scientific articles[2].
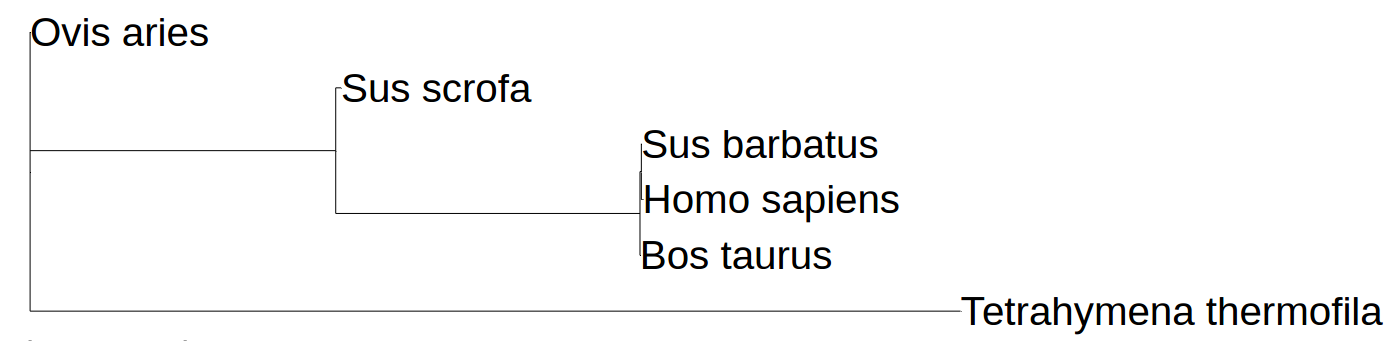
We made another 3D alignment to allow us to consider chains A and B of tubulin as similar. The results were quite peculiar. As you can see, the structures of A and B chain have no significant differences. There are a few noteable, though, which were marked with red or navy colors. This allowed us to consider both A and B chains identical. However, that gave us a few questions, e.g., how did this protein evolve and is it possible to perform a phylogenetic reconstruction with it. And it appears that there are some serious problems: the structure of tubulin was found for few species, and, as we managed to figure out, tubulin appears to be a conservative protein; such proteind lack the content needed by phylogenetic reconstruction programmes. You can see our attempt on the picture below. It is quite obvious, for example, that phylogenetic reconstruction programme cannot find the difference between the vertebrates. Structurally human tubulin does not differ much from one of swine's. All that confirms the assumption of tubulin's conservatism.
Personal impact:
Arsenii Lem wrote applet scripts and this web-page, implemented the 3D-alignments, and made an attempt of phylogenetic reconstruction; found the ligands on PubChem and described them.
Alexey Popov wrote the text of this article and found other articles to confirm the assumptions made here.
Sources:
- A. Lehninger – Principles of Biochemistry
- D. Sackett, J. Knutsonf, J. Wolff – Hydrophobic Surfaces of Tubulin Probed by Time-resolved and Steady-state Fluorescence of Nile Red, J. Biol. Chem.
- J. Cueva, J. Hsin, K. Huang, M. Goodman – Posttranslational Acetylation of α-Tubulin Constrains Protofilament Number in Native Microtubules
- D. Sept – Microtubule Polymerization: One Step at a Time, Current Biology
- Walker RA, O'Brien ET, Pryer NK, Soboeiro MF, Voter WA, et al. – Dynamic instability of individual microtubules analysed by video light microscopy: rate constants and transition frequencies., J. Cell Biol.
© Arsenii Loginovskii, Alexey Popov, 2016
