| Название | Мнемоника |
|---|---|
| Bordetella pertussis | BORPE |
| Burkholderia mallei | BURMA |
| Escherichia coli | ECOLI |
| Neisseria meningitidis | NEIMA |
| Thiobacillus denitrificans | THIDA |
| Paracoccus denitrificans | PARDP |
| Pseudomonas aeruginosa | PSEAE |
1. Поиск гомологов blastp
Алгоритм поиска гомлогов белка CLPX_ECOLI в протеомах выбранных бактерий:
- Создание fasta-файла ecoli_clpx.fasta с последовательностью CLPX_ECOLI
- Копирование протеомов выбранных бактерий в рабочую директорию
- Создание fasta-файла с протеомами всех выбранных бактерий:
cat * > proteomes.fasta
- Создание БД для поиска blastp из протеомов выбранных бактерий:
makeblastdb -dbtype prot -in proteomes.fasta -out clpx_db - Поиск гомологов по запросу ecoli_clpx.fasta в созданной ДБ с порогом e-value 0.001 c выдачей результатов в виде таблицы homologs.txt:
blastp -query ecoli_clpx.fasta -db clpx_db -evalue 0.001 -out homologs.txt
Выдача blastp (cсылка на полный файл)
Score E
Sequences producing significant alignments: (Bits) Value
sp|P0A6H1|CLPX_ECOLI ATP-dependent Clp protease ATP-binding subun... 860 0.0
sp|Q9I2U0|CLPX_PSEAE ATP-dependent Clp protease ATP-binding subun... 654 0.0
sp|Q3SI99|CLPX_THIDA ATP-dependent Clp protease ATP-binding subun... 642 0.0
sp|Q62JK8|CLPX_BURMA ATP-dependent Clp protease ATP-binding subun... 617 0.0
sp|Q7VXI6|CLPX_BORPE ATP-dependent Clp protease ATP-binding subun... 613 0.0
sp|A1B1H7|CLPX_PARDP ATP-dependent Clp protease ATP-binding subun... 580 0.0
sp|Q9JTX8|CLPX_NEIMA ATP-dependent Clp protease ATP-binding subun... 557 0.0
sp|A1B5T0|HSLU_PARDP ATP-dependent protease ATPase subunit HslU O... 103 3e-24
sp|Q9HUC5|HSLU_PSEAE ATP-dependent protease ATPase subunit HslU O... 95.9 1e-21
sp|P0A6H5|HSLU_ECOLI ATP-dependent protease ATPase subunit HslU O... 93.6 7e-21
sp|Q7VUJ9|HSLU_BORPE ATP-dependent protease ATPase subunit HslU O... 93.6 7e-21
tr|Q3SFW1|Q3SFW1_THIDA ATP-dependent protease ATPase subunit HslU... 86.7 2e-18
sp|Q62F00|HSLU_BURMA ATP-dependent protease ATPase subunit HslU O... 82.4 4e-17
tr|A1B8N4|A1B8N4_PARDP ATP-dependent Clp protease, ATP-binding su... 50.1 1e-06
sp|P0AAI3|FTSH_ECOLI ATP-dependent zinc metalloprotease FtsH OS=E... 46.2 2e-05
tr|A1AZV8|A1AZV8_PARDP ATP-dependent zinc metalloprotease FtsH OS... 44.3 8e-05
tr|A0A0H2WJ72|A0A0H2WJ72_BURMA ATP-dependent zinc metalloprotease... 43.9 9e-05
tr|Q3SJR4|Q3SJR4_THIDA ATP-dependent zinc metalloprotease FtsH OS... 43.5 1e-04
sp|A1AZW1|RUVB_PARDP Holliday junction branch migration complex s... 43.1 1e-04
tr|Q9HV48|Q9HV48_PSEAE ATP-dependent zinc metalloprotease FtsH OS... 43.5 2e-04
sp|Q9JUB0|RUVB_NEIMA Holliday junction branch migration complex s... 42.7 2e-04
sp|P0ABH9|CLPA_ECOLI ATP-dependent Clp protease ATP-binding subun... 43.1 2e-04
tr|Q3SJH1|Q3SJH1_THIDA ATP-dependent Clp protease, ATP-binding su... 42.7 3e-04
tr|A1BBJ2|A1BBJ2_PARDP ATP-dependent zinc metalloprotease FtsH OS... 41.6 6e-04
tr|A1AY35|A1AY35_PARDP Chaperone protein ClpB OS=Paracoccus denit... 41.6 7e-04
tr|A0A0U1RJ22|A0A0U1RJ22_NEIMA Replication-associated recombinati... 40.8 9e-04
2. Реконструкция и визуализация
Ссфлка на fasta-файл с последовательностями находокFasta-файл с последовательностями находок был загружен на NGPhylogeny.fr, где с помощью программы FastME было дерево на основе выравнивания MAFFT. Параметры: "Gamma distributed rates across sites" — No, "Starting tree" — BIONJ, "No refinement", 100 бутстреп реплик
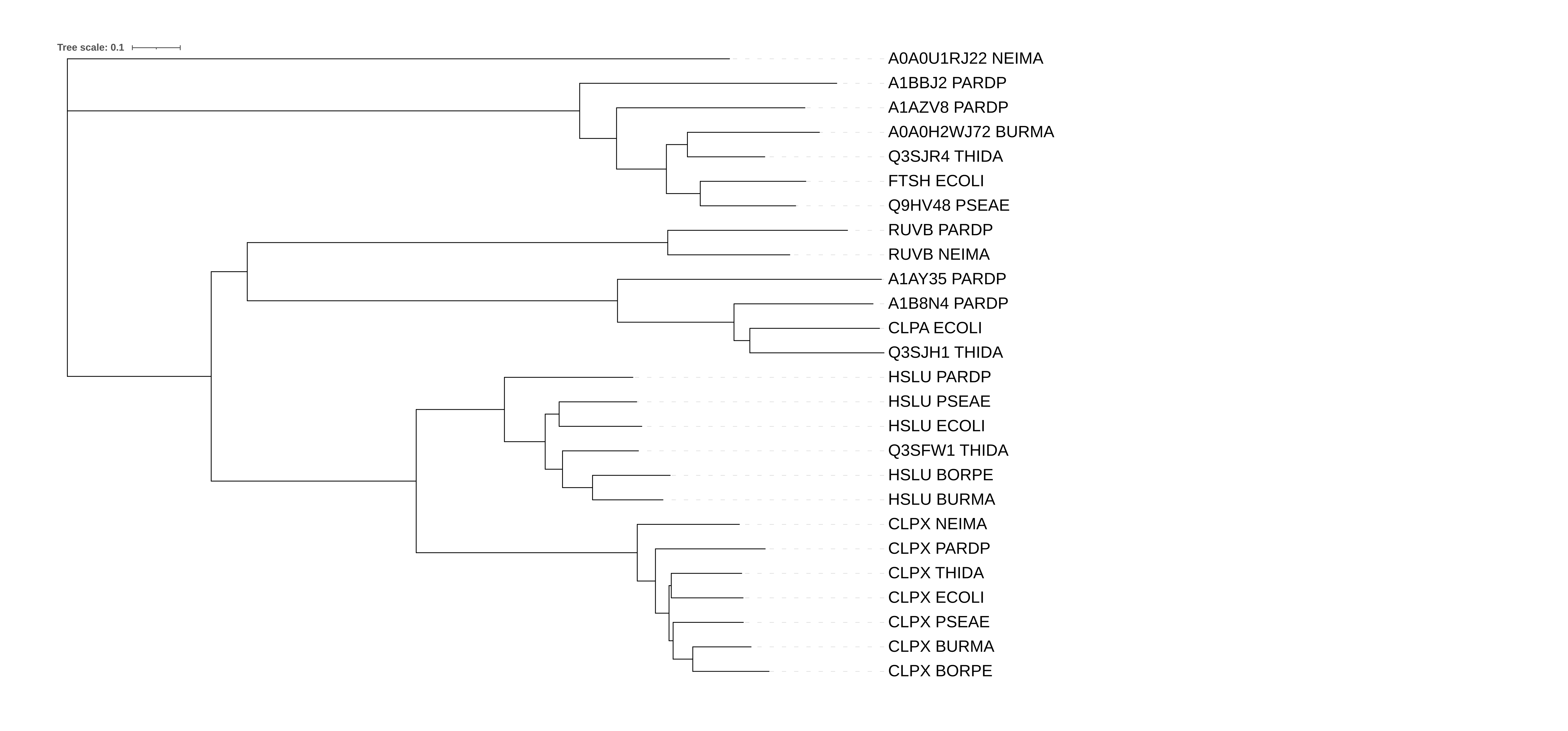
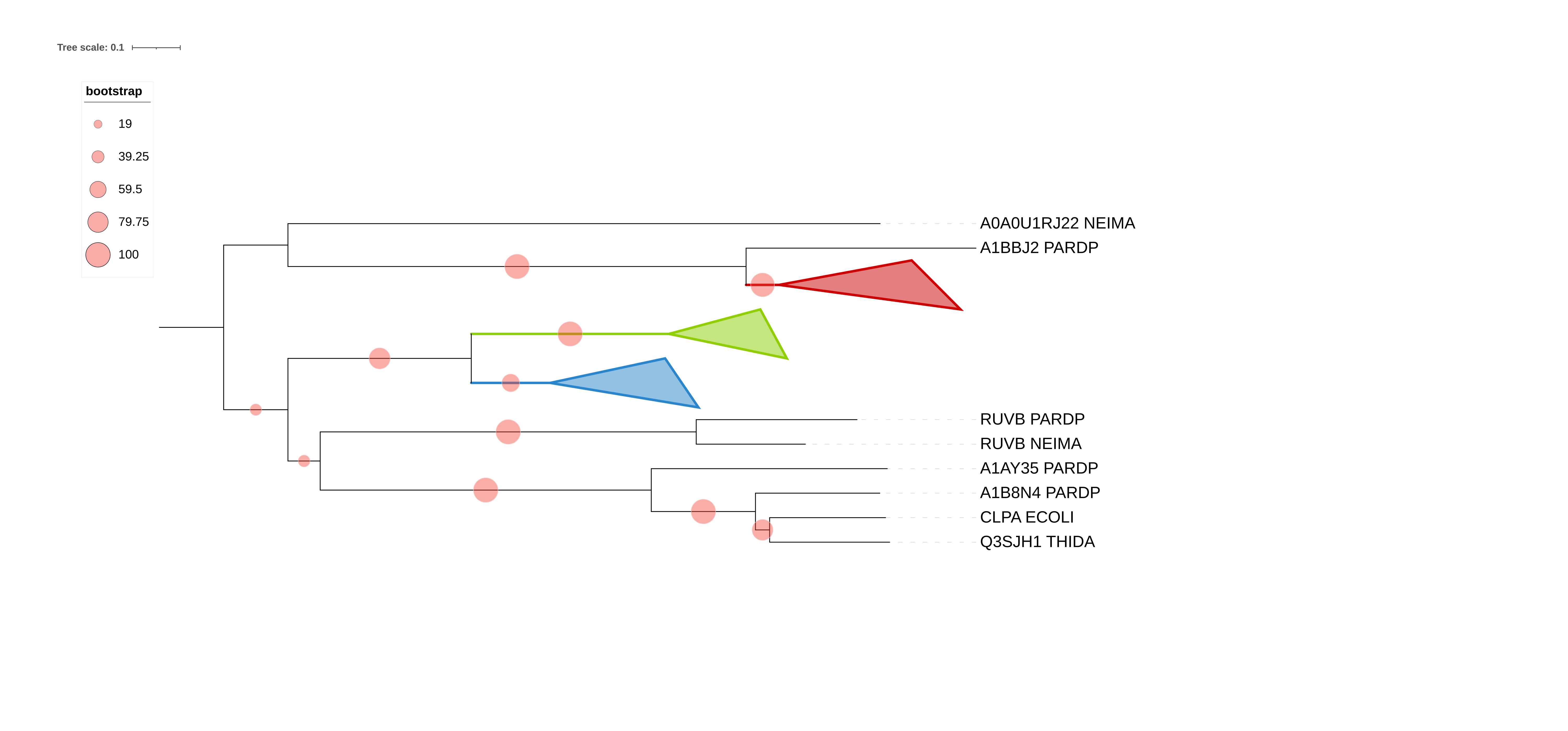
Ортологи:CLPX_BORPE и CLPX_BURMA, HSLU_PSEAE и HSLU_ECOLI, CLPX_ECOLI и CLPX_THIDA
Паралоги:CLPX_ECOLI и CLPA_ECOLI, Q3SFW1_THIDA и Q3SJH1_THIDA, Q9HV48_PSEAE и HSLU_PSEAE.
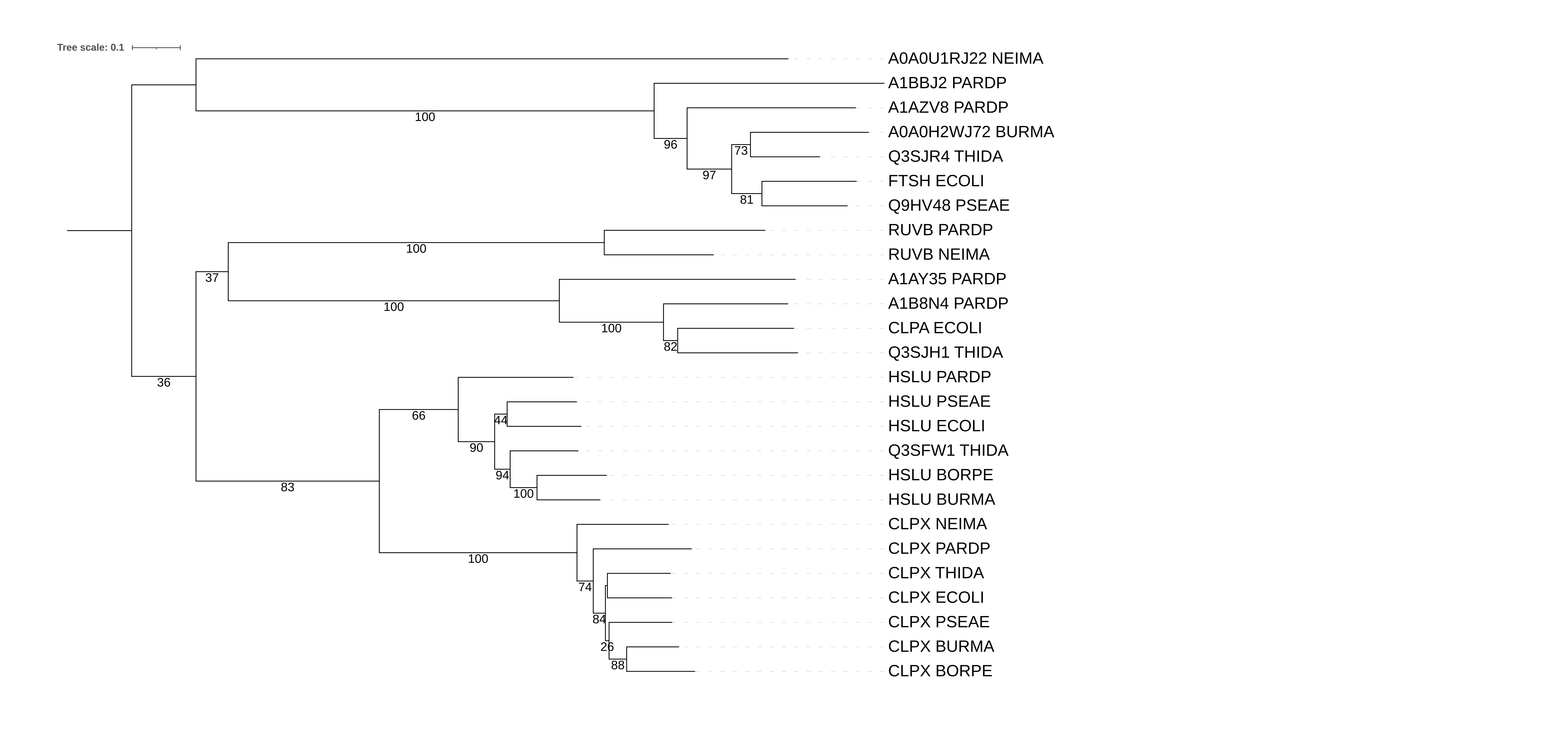
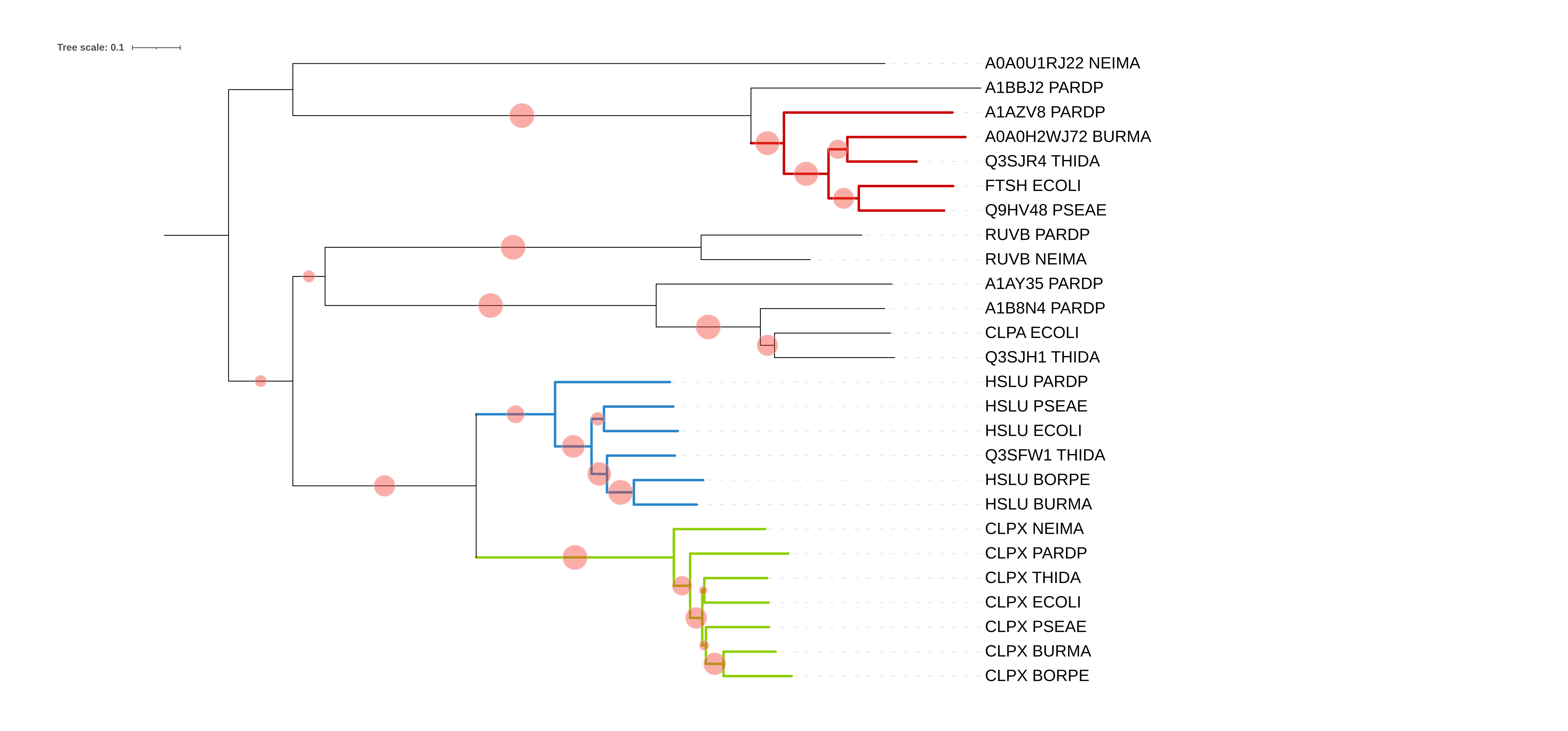
- красным - АТФ-зависимая цинковая металлопротеаза (ATP-dependent zinc metalloprotease). В эту группу входят белки пяти выбранных бактерий, не вошли - NEIMA, BORPE. Топология совпадает с исходных деревом.
- синим - АТФ-зависимая субъединица АТФ-зависимой протеазы HslU (ATP-dependent protease ATPase subunit HslU). В эту группу входят белки шести выбранных бактерий, не вошла - NEIMA. Топология совпадает с исходных деревом.
- зеленым - АТФ-связывающая субъединица АТФ-зависимой Clp-протеазы (ATP-dependent Clp protease ATP-binding subunit). В эту группу входят белки всех семи выбранных бактерий. Наблюдается несовпадение с топологией исходного дерева: разрушилась группа betaproteobacteria - в нее правильно вошли BORPE и BURMA, но THIDA стоит далеко от них. THIDA, ECOLI, PSEAE, BURMA и BORPE входят в одну кладу, то есть получается, что группа betaproteobacteria перемешана с gammaproteobacteria.