1.Introduction
This page shows the information about protein complex between human phosphopantenil transferase and acyl-carrier protein (ACP) [1]. Experimental data has gotten using X-Ray difraction. Acyl-carrier protein forms a part of fatty acid synthetase. It is a multi-domeins proteins consisted of 6 subunits (ACP is not counted). Reactions of fatty acids' synthesis are shown on picture 1.
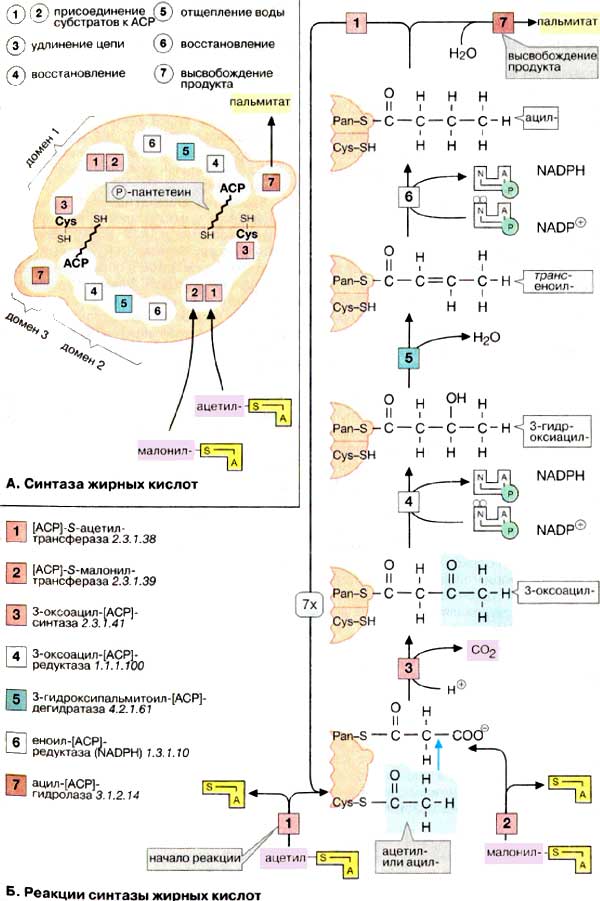
2. Description of proteins included in complex 2CG5
| L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | Acyl-carrier protein (as a part of fatty acid synthetase ) | |
| Short name | AASD-PPT | ACP |
| Gene name | AASDHPPT | Fasn |
| PDB ID | 2BYD | 2PNG |
| UniProtKB AC | Q9NRN7 | P12785 |
| Molecular function | Transferase | Transferase |
| Catalytic activity | CoA-[4'-phosphopantetheine] + apo-[acyl-carrier-protein] = adenosine 3',5'-bisphosphate + holo-[acyl-carrier-protein]. | Fatty acids synthesis (pic.1) |
| Ligands | Magnesium, CoA, Br | B5 |
| Cellular localisation | Cytosol | Cytosol |
Phosphopantetheinyl transferase - it is an enzyme, capable to transfer 4'-phosphopantetheine residue of CoA to conserved serine-2156 residue of APB. It is also able to phosphopantetheinylate peptid- and acyl-carrier procariotic proteins. Moreover, this enzyme takes part in lysine catabolism.[3].
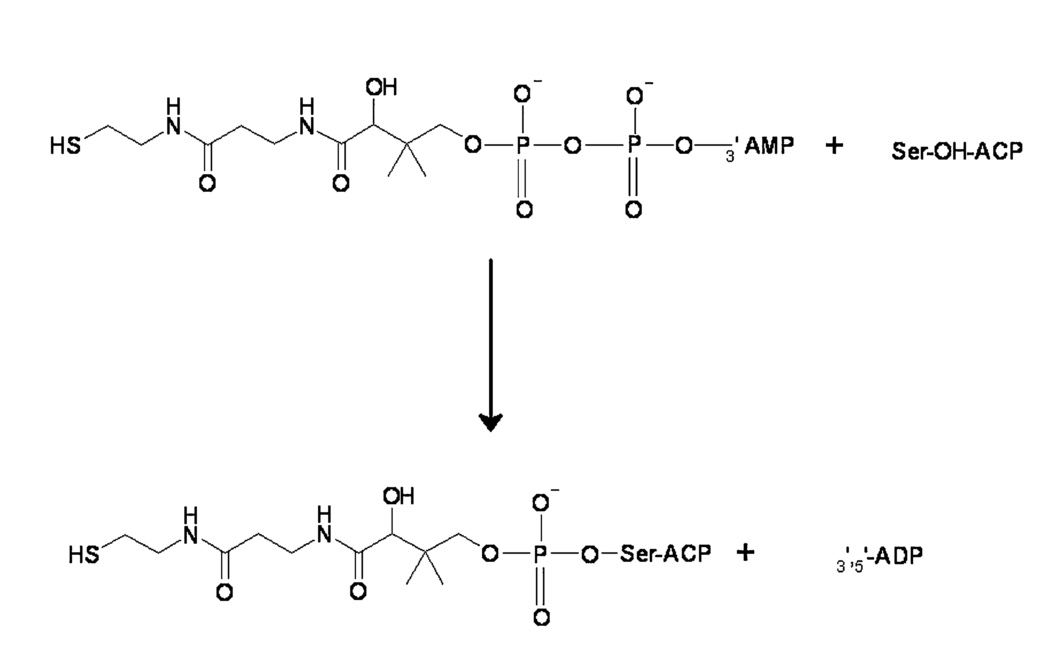
Acyl-carrier protein is a important component in fatty-acids and polycetide (secondary metabolite [5]) biosynthesis. It is synthesized in inactive form and activated throw joining 4'-phosphopantetheine. Phosphopantetheine have two vital functions: joininf of synthesing intermediate to fatty-acid synthase throw energy-rich link and because of flexibility of its chain allows to bring together intermediates and enzyme active center [6].
3. Protein-protein contcts
| Generalconfiguration | Script |
| Cores | Script |
| Polarinteractions | Script |
| Hydrogenbond | Script |
| Aromatic | Script |
| Resume | |
| Save PNG | |
| Press 'Resume' to switch pictures | |
Hydrophobic interactions . There are lots of alpha-helixes in either proteins so we can suppose that there are lots of hydrophobic interactions between them.
Using server [7] we identified hydrophobic cores of proteins. Using within-command (average distance between two covalent not-binded atoms we know from the previous task-4 angstrems [8]) we studied that only 4 from overall 10 cores takes part in protein-protein contacts. If you pull 'Cores'-buttom, you will see a general view of these cores (red-transferase, black-ACP, another colors-different cores). If you pull 'Resume'-button, you will se a general scheme of hydrophobic interactions (List of aminoacids, takes part in it ACP:[ALA]2130 [ILE]2134 [ILE]2137 [LEU]2152 [ALA]2156 [LEU]2157 [VAL]2160 [GLU]2161 [THR]2165 [VAL]2176 Transferase: [PHE]61 [VAL]62 [PHE]63 [GLN]122 [PHE]154 [ILE]157 [TYR]184 [TRP]187 [LEU]201). Transferase displayed in grey, ACP in green, aminoacids from transferase, taking part in hydrophobic interactions, in pink, from ACP in blue. It is clearly seen that hydrophobic contacts are aggregated in two regions, keeping ACP from two sides. One of them is situated between 'twist-region' of secondary structure (pull 'Resume'-button) and one of them between alpha-helixes (pull 'Resume'-button). We suppose that hydrophobic interactions help to orientate correct the movements of two proteins relative to one another and keeping the right position for some time, necessary for reaction.
Polar interactions. As a rule, it is different to draw a line between polar interactions and hydrogen bonds, so we mean that there are bonds between positive and negative charged residues not lonfer than 5 angstrom.
We have found only three polar bonds ([GLU]2161:B -[Arg]148:A, [Thr]2165:B -[Arg]148:A и [Asp]2151:B-[Gln]60:A). Pull 'Polar interactions'-button to see a genera view, where ACP is diplayed in green, transferase in grey, negative charged residues in blue, positive charged in pink. Press 'Resume'-button to see them closer. On picture 4 you can see a snapshot of ASP2151-GLN60 where high atoms of aminoacids are marked.
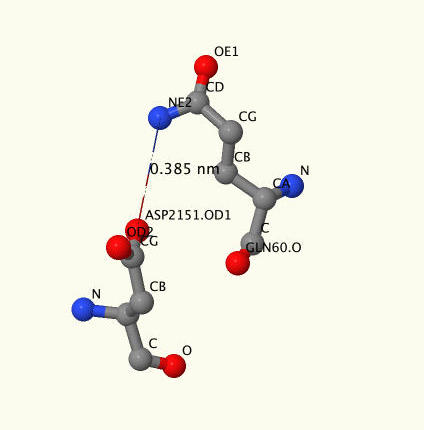
Hydrogen bonds.There is only one hydrogen bond between oxygen of ASP2151 and nitrogen of VAL62. For visualisation press 'Hydrogen bond'-buttom. This link is unusual because of the fact, that it is not between lateral-lateral residues but between lateral residue and backbone.
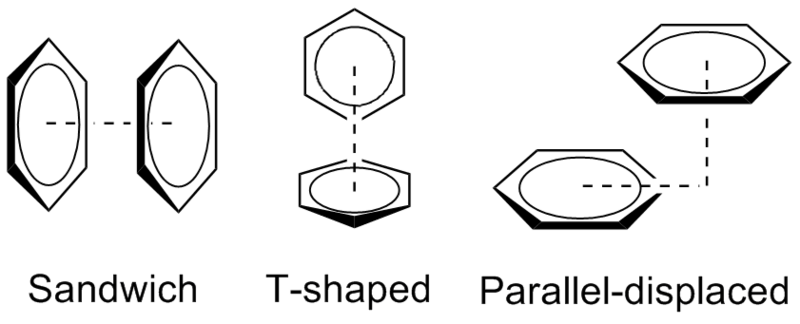
Stacking-interactions. Stacking-interaction - it is attractive, noncovalent interaction between aromatic rings, containing pi bonds [9]. Using within command we found aromatic (phe, trp, tyr, his) aminoacids, siteated on the contacts' broad between transferase and ACP (trp187, phe154, phe63, phe 61, his2133, tyr184). For visualisation press 'Aromatic'-buttom. As it seen, all of them are situated too far from each other to form a stacking-bond (1,26 nm and 1,09 nm).
However we found information about stacking berween aromatic aminoacids and nitrogen atoms of glu, arg and asp [10]. Authors of that article said that there are stacking-interactions no longer than 5 angstrom. Press 'Resume'-button to see PHE63-ARG65 and ARG2138-HIS2133. We assume that these interactions help to stabilize 'twist-structure' of alpha-helix. It can play role in pritein positioning (press 'Resume'-button to see general view).
4.Ligands in complex 2CG5, 2BYD, 2JBZ.
In this chapter we review protein-ligand contacts in 2CG5 complex, and, separately, in 2BYD and 2JBZ. There are 6 different ligands in total, their features are presented in Table 2.| 2BYD | Script |
| 2JBZ | Script |
| 2CG5 | Script |
| Resume | |
| Save PNG | |
| Press "Resume" to switch pictures | |
| Ligand | CoA | Br- | Ni2+ | K1+ | Mg2+ | PO43- |
| PDBID | 2CG5 and 2JBZ | 2BYD | 2CG5 | 2JBZ | 2CG5 and 2JBZ | 2CG5 |
| IUPAC | S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] ethanethioate | Bromide | Nickel(2+) | Potassium(1+) | Magnesium(2+) | Phosphate |
| Chemical formula | 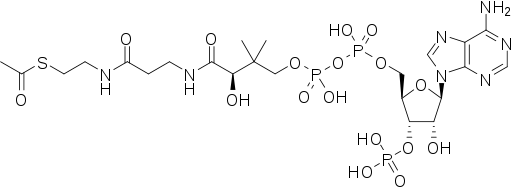 | 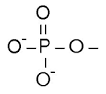 | ||||
| Gross formula | C23H38N7O17P3S | Br | Ni | K | Mg | PO4 |
| Molar mass | 809,57 | 79.90 | 58.69 | 39.10 | 24.30 | 94,97 |
| PubChem | 444493 | 259 | 934 | 813 | 888 | 1061 |
5.Conclusions
Taking everything into account we can say that our proteins connected to each other with hydrophobic interactions and polar interactions do not play a main role in this process. Opposite, polar small ligands positioned in proteins by binding with polar residues of aminoacids.6.References
[1] http://www.rcsb.org/pdb/explore/explore.do?structureId=2CG5
[2] Наглядная биохимия. Кольман Я., Рём К.-Г. М.: 2000. - 469 с
[3] Joshi A.K. et al. Cloning, Expression, and Characterization of a Human 4’-Phosphopantetheinyl Transferase with Broad Substrate Specificity // J. Biol. Chem. 2003. Vol. 278, № 35. P. 33142–33149.
[4] Bunkoczi G. et al. Mechanism and Substrate Recognition of Human Holo ACP Synthase // Chem. Biol. 2007. Vol. 14, № 11. P. 1243–1253.
[5] https://ru.wikipedia.org/wiki/Поликетиды
[6] https://meshb.nlm.nih.gov/#/record/ui?name=Acyl%20Carrier%20Protein
[7] http://mouse.belozersky.msu.ru/npidb/cgi-bin/hftri.pl
[8] http://kodomo.fbb.msu.ru/~azbukinanadezda/term2/pr2/pr2.html
[9] https://en.wikipedia.org/wiki/Stacking_(chemistry)
[10] Flocco M.M., Mowbray S.L. Planar stacking interactions of arginine and aromatic side-chains in proteins. // Journal of molecular biology. 1994. Vol. 235. P. 709–717.