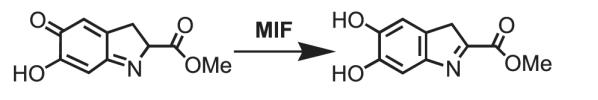
1. MIF tautomerizes dopachrome methyl esters [4]
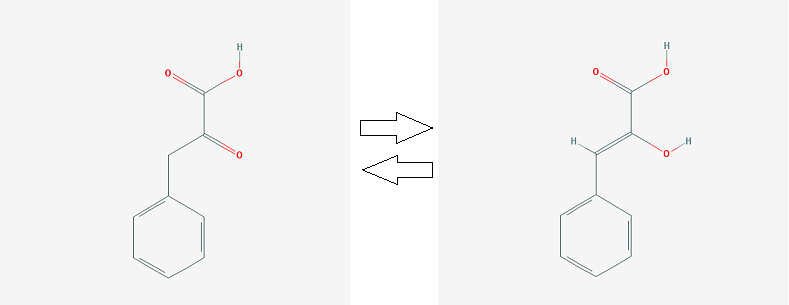
2. keto-phenylpyruvate <=> enol-phenylpyruvate (MIF catalyzes reaction)
The contact atlas of Macrophage Migration Inhibitory Factor in Complex with a Naphthyridinone Inhibitor (pdb id = 6B1K)
Introduction
Macrophage migration inhibitory factor (MIF) is a pro-inflammatory cytokine, which plays a key role in innate immunity to bacterial patogens and inflammation (it binds to CD74 and trigger an immune response), counteracts the anti-inflammatory activity of glucocorticoids, has phenylpyruvate tautomerase and dopachrome tautomerase activity [1].

1. MIF tautomerizes dopachrome methyl esters [4]

2. keto-phenylpyruvate <=> enol-phenylpyruvate (MIF catalyzes reaction)
MIF’s expression is shown in endothelial cells, macrophages, T-cells. Elevated levels of MIF is observed in some neurological diseases (rheumatoid arthritis, atherosclerosis, Alzheimer's disease), its role in cancer development is suggested [2]. Identifier of the protein in UniProt database: P14174. The protein consists of 3 identical subunits, its total mass is about 38,5 kDa. Each of its subunits composed of 114 residues contain two antiparallel alpha helices and a four-stranded beta sheet.
Ligands
Considered protein is connected with eight ligands. The main of them are three molecules of C9G, which inhibit an enzymatic activity of the protein. Sulfate ions and glycerol were in experimental solution (2,1 M ammonium sulfate is one of the components described) and don’t have any function.
 |
 |
 |
Protein-ligand contacts
All contacts were found using the command "within"
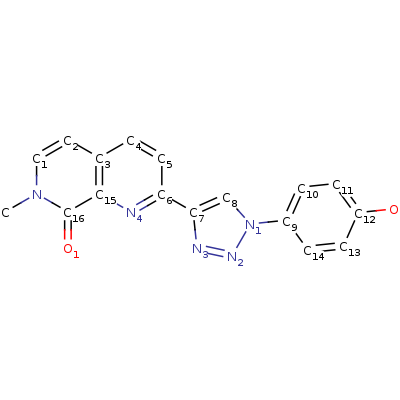
C9G
2-[1-(4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]-7-methyl-1,7-naphthyridin-8(7H)-one (C9G) inhibits MIF’s tautomerase activity through building in protein’s active center: according to the article [2] hydrogen bonds occur between Asn97 and the phenolic hydroxyl group of the inhibitor, the backbone NH of Ile64 and N2 of the triazole, and the ammonium group of Lys32 with N3 of the triazole, the quinoline nitrogen, and the carbonyl oxygen of Ile64. There are also an aryl?aryl interactions with Tyr36, Tyr95, and Phe113 (stacking) and hydrophobic interactions shown on pdb.
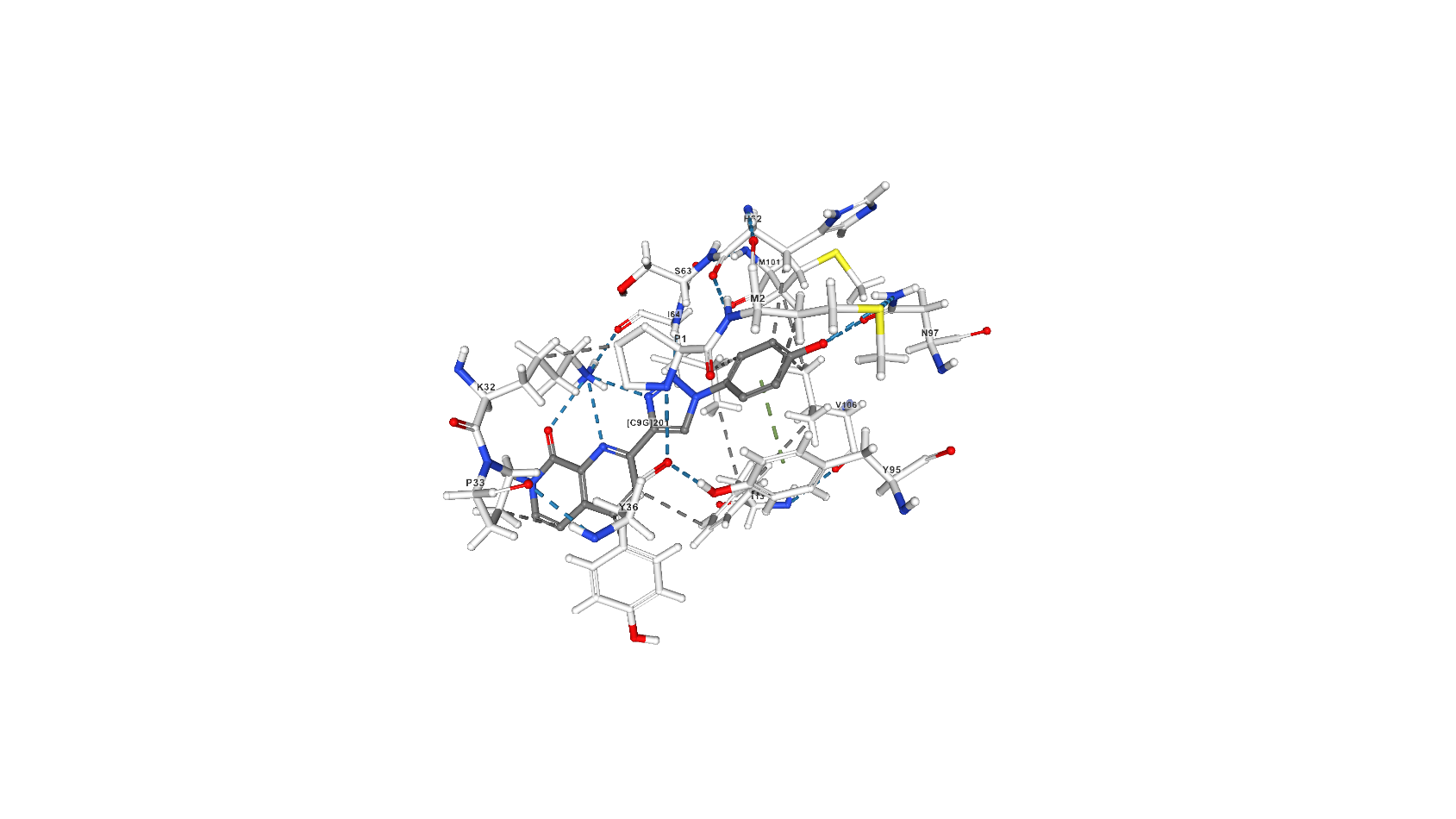
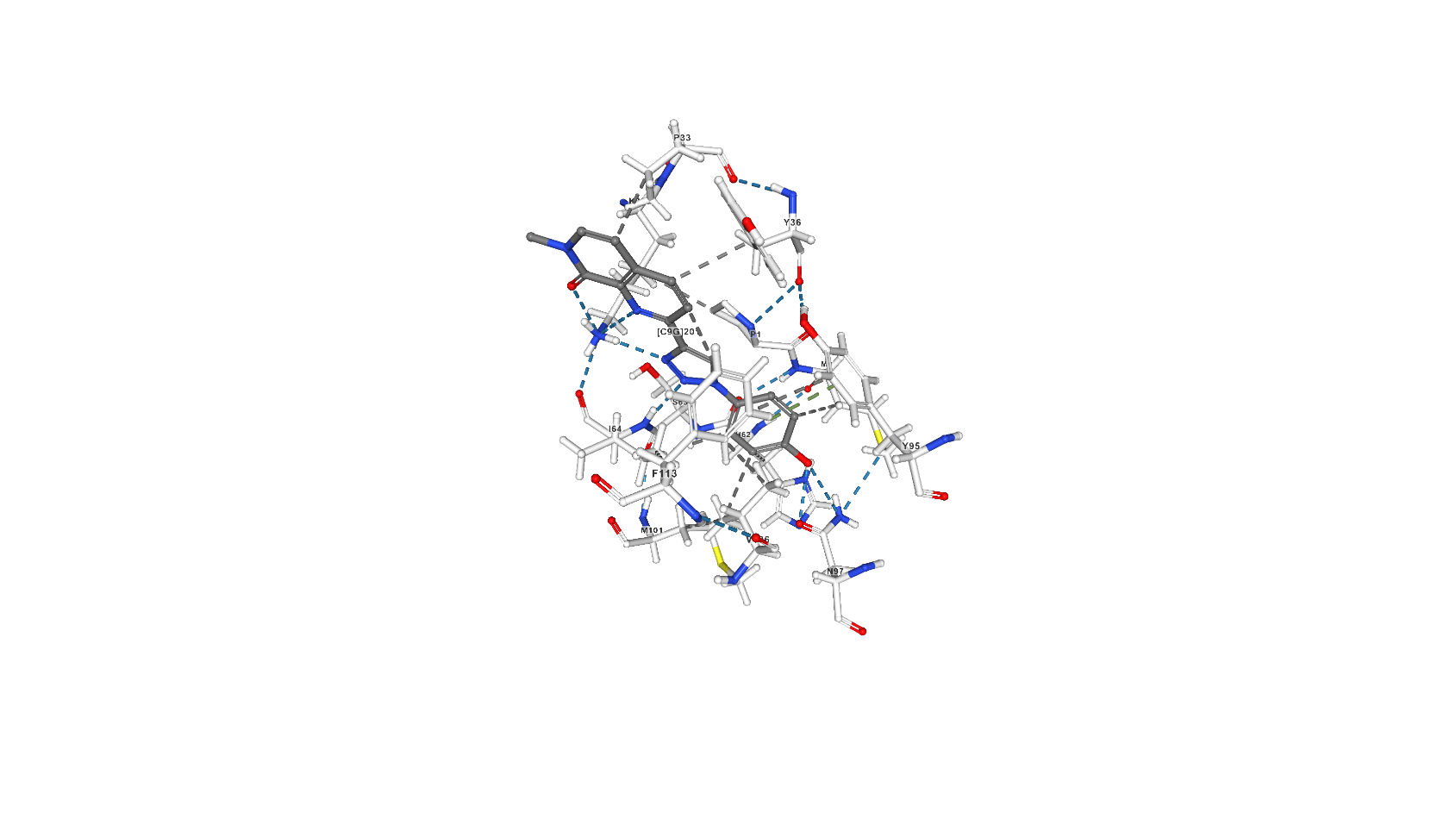
Screenshots with C9G from pdb
Glycerol (GOL)
Glycerol makes 2 hydrogen bonds with Asp92 of chain A. Atoms of glycerol involved in this interaction are [ASP]92:A.O and [ASP]92:A.OD2. Also, hydrogen bonds occur between glycerol and water (4 bonds). Another type of bonds aren’t found.
SO4-ligand
The protein is bound to 4 sulfate-ions. The interaction based only on hydrogen bonds. [SO4]202:A.O1 and [SO4]202:A.O4 is bound to water molecules and [SO4]202:A.O2 is bound to [ALA]70:A.H , [SO4]202:A.O3 is bound to 2 glutamine’s atoms :[GLN]71:A.H and [GLN]71:A.HE21.
Applet 1
Protein-protein contacts
There are no covalent bonds between MIF’s subunits.
Hydrogen bonds
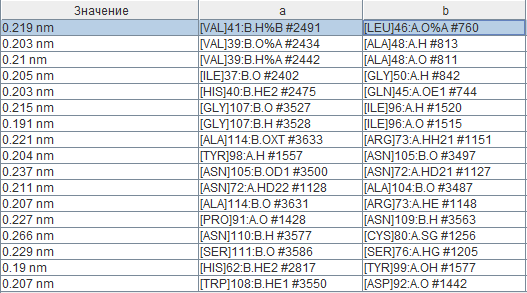
For the current moment 17 hydrogen bonds are found between chains a and b. As it can be seen from the image of the secondary structure, some of them are created through 2 “mixed” beta-sheets, where some strands belong to one chain and some strands to another one. In all pairs of chains interactions are similar so they weren’t considered. All residues which are not more than 4 Angstroms far from another chain were found using the command “within”. Lengths of hydrogen bonds are represented in the table (names of columns aren’t names of chains). The average length of bond is 0,214 nm or 2,14 Angstroms, which is less than usually shown in alpha-helix and beta-sheet (page with practicum 1). The shortest bond (0,19 nm) are found between Histidine (HE2) and Tyrosine (OH-group oxygen) residues, which is possibly explained by their basic and weak acid properties respectively. Interactions between 45-50 residues of one chain (beta3-strand) and 37-42 of another one (beta2-strand) is described in literature [3] as one of the key interactions for the considered protein which takes part in maintaining its quaternary structure.
Hydrophobic interactions
It is well-noticed in the picture with bright-colored hydrophilic amino acids ands and pale-colored hydrophobic ones that external-oriented parts of the protein are more-likely to be hydrophilic and more internal parts are hydrophobic. Atoms of hydrophobic residues account for 60% of total. As an example of hydrophobic core Tyr95 of chain A and its surroundings were shown. Hydrophobic interaction begins to act beyond 1 Angstrom. At the distance more than 5 Angstroms the ambit of the central residue is so dense that it becomes poorly discernible behind its neighbors. According to the measurements done, an average distance between neighbor not bound covalently atoms is approximately 3,5 Angstroms. The diameter of the water molecule neglecting hydrogen is 2,8 Angstroms, which forbids it to build in the gap: 0,35 Angstroms (0,5*(3,5-2,8)) is less than a covalent link length, repulsion forces between atoms are too strong so they push out an extra molecule.
Another example of hydrophobic interaction is found in the C-terminal region of the protein (residues 105-114, namely [VAL]106, [GLY]107, [TRP]108, [PHE]113). It was described that deletion of these residues perturbs the tertiary structure of the protein and leads to loss of its enzymatic activity, reduction in macrophage activating properties but has no influence on its receptor binding properties[3].
Salt bridges (ionic interactions)
Ten salt bridges were found. The bond between Arginine and Glutamic acid residues is represented in most cases. The interaction was shown using the command "measure".
Applet 2
Author contributions:
Kirill Tsyganov selected the protein, described its physico-chemical properties and protein-ligand interactions for glycerol and sulfate ion, found salt bridges, made an applet for ligands. Darya Bykova described hydrophobic interactions, hydrogen bonds and protein-inhibitor interactions and also designed the html-page.
References:
1. http://www.uniprot.org/uniprot/P14174
2. Dawson, T.K., Dziedzic, P., Robertson, M.J., Cisneros, J.A., Krimmer, S.G., Newton, A.S., Tirado-Rives, J., Jorgensen, W.L. Adding a Hydrogen Bond May Not Help: Naphthyridinone vs Quinoline Inhibitors of Macrophage Migration Inhibitory Factor. ACS Med Chem Lett, 2017, 8: 1287-1291
3. Farah El-Turk, Bruno Fauvet, Amer Ashrafi, Hajer Ouertatani-Sakouhi, Min-Kyu Cho, Marilisa Neri, Michele Cascella, Ursula Rothlisberger, Florence Pojer, Markus Zweckstetter, Hilal Lashuel. Characterization of Molecular Determinants of the Conformational Stability of Macrophage Migration Inhibitory Factor: Leucine 46 Hydrophobic Pocket. PLoS One. 2012; 7(9): e45024.
4. Crichlow GV1, Cheng KF, Dabideen D, Ochani M, Aljabari B, Pavlov VA, Miller EJ, Lolis E, Al-Abed Y. Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor. J Biol Chem. 2007 Aug 10;282(32):23089-95. Epub 2007 May 25.
5. https://en.wikipedia.org/wiki/Macrophage_migration_inhibitory_factor
© Áûêîâà Äàøà, 2018