Mycoplana bullata (ramosa) is a heterotrophic bacterium which habitat is regularly alkalescent. [1]
Polyamines are essential aliphatic polycations that bind to nucleic acids and accordingly are involved in a variety of cellular processes. Polyamine function can be regulated by acetylation and deacetylation, just as histone function can be regulated by lysine acetylation and deacetylation. Acetylpolyamine amidohydrolase (APAH) from Mycoplana ramosa is a zinc-dependent polyamine deacetylase that shares approximately 20% amino acid sequence identity with human histone deacetylases. [2]
7-Amino-N-hydroxyheptanamide | |||
IUPAC |
[7-(hydroxyamino)-7-oxoheptyl]azanium |
||
Molecular formula |
C7H17N2O2+ |
||
Molar mass |
161.225 g/mol |
||
| PubChem | 91824280 |
||
It’s a potent inhibitor of histone deacetylases and induces cell cycle arrest and apoptosis in several human cancer cell lines. [3]
Glycerol |
|
IUPAC |
propane-1,2,3-triol |
Molecular formula |
C3H8O3 |
Molar mass |
92.094 g/mol |
| PubChem | 753 |
Potassium cation | |||
IUPAC |
potassium(1+) |
||
Molecular formula |
K+ |
||
Molar mass |
38.964 g/mol |
||
| PubChem | 813 |
||
Zinc cation | |||
IUPAC |
zinc(2+) |
||
Molecular formula |
Zn+2 |
||
Molar mass |
65.38 g/mol |
||
| PubChem | 32051 |
||
Azanium | |||
IUPAC |
azanium |
||
Molecular formula |
H4N+ |
||
Molar mass |
18.034 g/mol |
||
| PubChem | 223 |
||
Ligands such as K+ and Zn2+ cations form ionic and dative type interactions with nearby negative charged groups of different residues. Azanium cation forms hydrogen bonds with the closest atoms of oxygen in the vicinity. (those hbonds cannot be shown in JMol). [19]
The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water molecules. [16] In the case of protein folding, the hydrophobic effect is important to understanding the structure of proteins that have hydrophobic amino acids (such as glycine, alanine, valine, leucine, isoleucine, phenylalanine, tryptophan and methionine) clustered together within the protein. Structures of water-soluble proteins have a hydrophobic core in which side chains are buried from water, which stabilizes the folded state. [17] PHE27 was picked in the capacity of “MyResidue”. Surrounding atoms which are 4 Å away from residue almost cover it whole. To determine the typical distance between neighboring noncovalent bound atoms several measurements were made and so it’s accounted for 4.32 Å on the average. The mean distance between van-der-Waals radii is 0.62 Å. Oxygen atom with the radius of 1.4 Å won’t fit between 2 neighboring atoms, let alone the water molecule. [18]
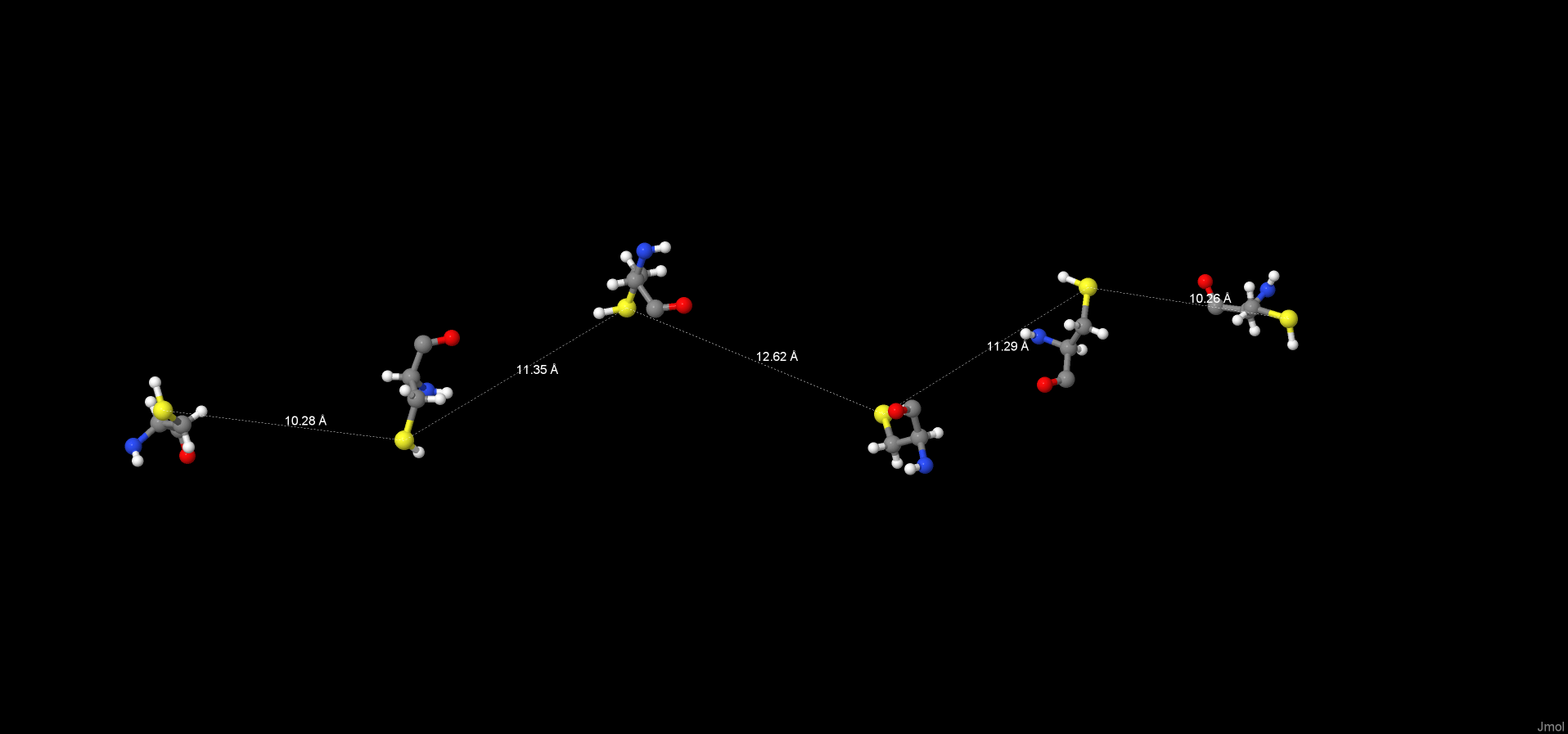
Disulphide bond is often formed between two residues of cysteine inside one of polypeptide chains or between two of those therefore stabilizing the protein molecule. [4] ] The search of disulphide bridges was done manually by measuring the distance between atoms of Sulphur from different cysteine residues. Due to the fact that the minimal measured distance is close to 10.26 Å the formation of this type of interactions is considered as impossible (the average disulphide bond length is 2.05 Å). [5]
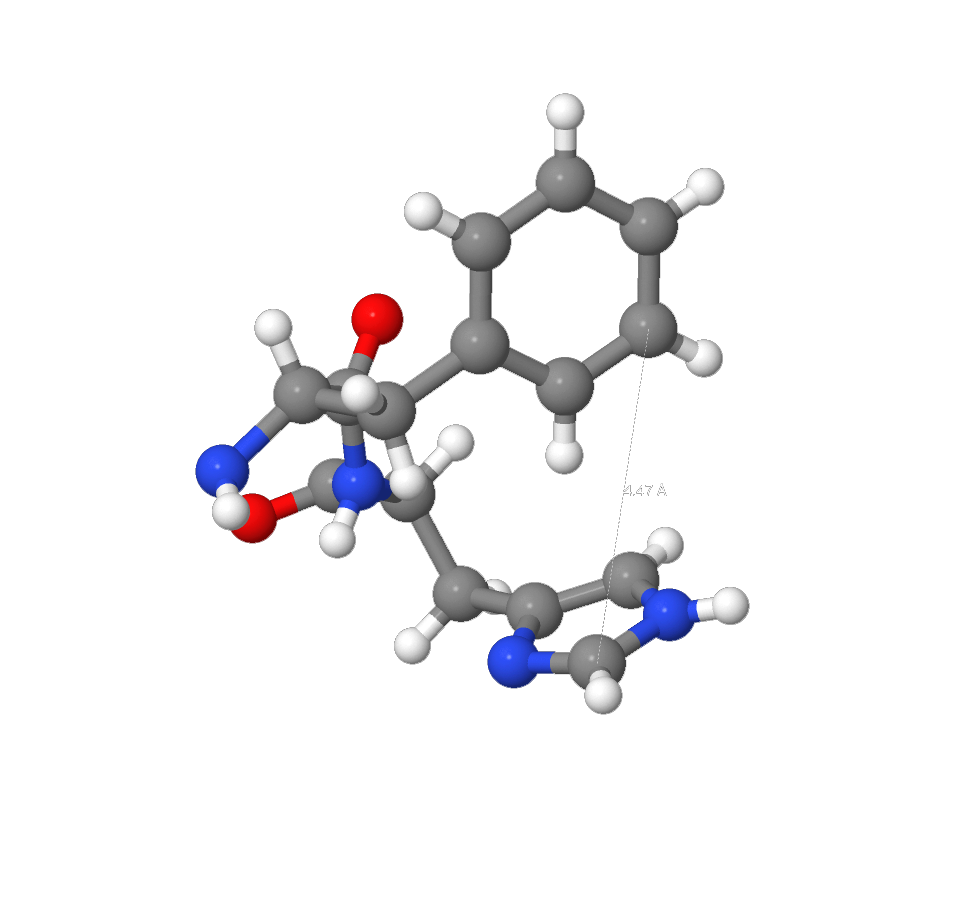
In organic chemistry aromatic interaction (or π-π interaction) is a noncovalent interaction between organic compounds which contain aromatic components. π-π interactions are caused by intermolecular overlaps of P atomic orbitals in π-systems which make them stronger as the result of increased amount of pi electrons. Other noncovalent interactions include Van-der-Waals forces, dipole/dipole interactions and interactions associated with charge transfer. Stacking interactions were found near atom of Zn by using Ligand view model from RCSB PDB site. (Although this interaction can be found by measuring the distance between aromatic structures which are parallel (decentralized parallel interaction) or perpendicular to each other by using JMol [6]. Additionally, the second of above-mentioned types of interactions was found between [HIS]196 and [PHE]197). Average length of interaction – 3.4 Å. [7]
Definition by IUPAC: The hydrogen bond is an attractive interaction between a hydrogen atom from a molecule or a molecular fragment X–H in which X is more electronegative than H, and an atom or a group of atoms in the same or a different molecule, in which there is evidence of bond formation. [8] The most common electronegative atoms in hydrogen bonds are F, O and N. As for biomolecules, the typical donor atoms are the oxygens in -OH (e.g. the sidechains of Ser, Thr, Tyr), HOH, and the nitrogen in -NH3+ (as in the sidechains of Lys, Arg) or -NH- (as in the main chain peptide bond, and the sidechains of Trp, His, Arg). The lone electron pairs on these same donors can serve as hbond acceptor sites. So can the atoms in =O or =N- groups but these latter cannot serve as donors. [9] Angle N-H-O in the hbond in the best case is close to 180° but in biomolecules the ± 45° error is possible and angles 130°-180° are considered as average. [10] Depending on the length between the donor and acceptor hbonds are categorized as weak (3.2-4.0 Å), moderate (2.5-3.2 Å) and strong (2.2-2.5 Å). Moderate hbonds are more often presented in biomolecules than the other ones. [10] [11] In 4zur protein hbond N(109ILE)-O(105ILE) in alpha-helix made from residues 104-112 in chain A was picked for analysis. The acceptor is an oxygen =O and donor is nitrogen –NH– from the main chains of corresponding residues. The distance between the donor and acceptor is 2.91 Å. Angle N-H-O equals 162.6°. And so, regarding the data, this particular hbond can be considered as standard for a biomolecule. To complete the research on hbonds with JMol the following commands were used – “calculate hbonds”; “restrict” (to restrict the needed residues); “measure” (for measuring the N-H-O angle and the length of a bond).
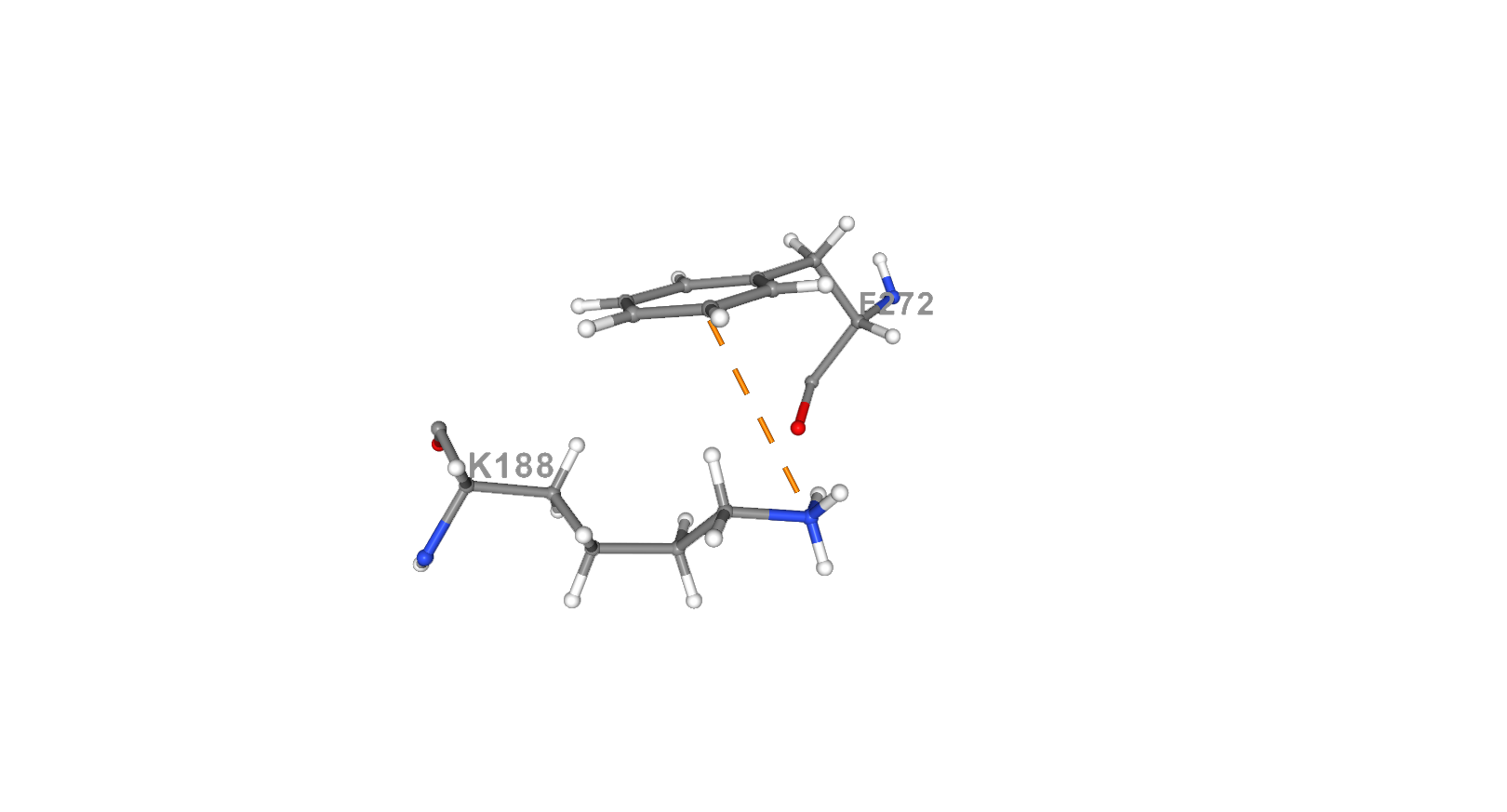
Cation parts of aminoacids can participate in polar interactions with aromatic rings of other residues if the distance between them is less than 6 Å. This kind of interactions called cation-pi. Aromatic rings have partially negative charge owing to the delocalized pi electrons. Cations such as sidechains of LYS and ARG align above the rings. Cation-pi interactions make a significant contribution to the overall stability of most proteins. [12] [13] In 4zur protein cation-pi interaction between [LYS]188:A.NZ- [PHE]272:A.CE2 was picked for analysis. The sidechain of lysine plays role of cation in this interaction while the aromatic ring of phenylalanine is a π-system with the negative charge. The distance between cation and π-system is 5 Å, which is less than 6 Å and so the interaction can be considered as effective. [13] For more efficient search of this interactions NGL viewer was used [14] as well as commands in Jmol such as “restrict” and “measure” which were used for restricting the needed residues and measuring the length of the interaction respectively.
Salt bridges can occur between positive charged groups of cysteine[CYS], arginine[ARG] or lysine[LYS] and negative charged groups of aspartic[ASP] and glutamic[GLU] acids. So that the salt bridges form on account of ionic interactions. The formation of salt bridges is possible when the bond length is no longer than 4 Å. [15] In 4zur protein the salt bridge between [ARG]32:B.NH2 и [GLU]28:B.O was picked for analysis. The distance between NH2 arginine group and COOH glutamic acid group stands for 3.53 Å so that the salt bridge can be considered as effective. JMol commands such as “restrict” and “measure” were used to restrict the needed residues and to measure the length of the salt bridge.
Amino-N-hydroxyheptanamide(APAH) is a ligand which connects to Zn2+ ions bridges the binuclear metal cluster (active site) while one of two Zn2+ ions is coordinated on is coordinated by the imidazole side chains of H158 and H159 as well as a water molecule. Consequently, Zn2+ is a keystone in the work of inhibitor as it unites all the components together. Commands such as “restrict” and “color bonds” were used to highlight the active site. [2]

[1] Mycoplana on Wikipedia [2] Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of their Complexes with Acetylpolyamine Amidohydrolase [3] Histone Deacetylase Inhibitor M344 Inhibits Cell Proliferation and Induces Apoptosis in Human THP-1 Leukemia Cells. [4] БИОЛОГИЧЕСКАЯ ХИМИЯ Березов Т.Т., Коровкин Б.Ф. [5] Formation and transfer of disulphide bonds in living cells [6] Rethinking the Term "Pi-Stacking" [7] Hunter & Sanders J.Am. Chem. Soc. 1990, v112, p 5525-5534 [8] Defining the hydrogen bond: An account (IUPAC Technical Report) [9] Hydrogen bonds on Proteopedia [10] Professor David A. Evans group seminar on Noncovalent interactions: An Introduction to Hydrogen Bonding (2009) [11] Jeffrey, George A., An introduction to hydrogen bonding, Oxford University Press, 1997 [12] Cation-pi interactions on Proteopedia [13] Cation-pi interactions in structural biology. [14] NGL Viewer [15] Salt bridges on Wikipedia [16] Chandler D (2005). "Interfaces and the driving force of hydrophobic assembly" [17] (Pace CN, Shirley BA, McNutt M, Gajiwala K (1 January 1996). "Forces contributing to the conformational stability of proteins" [18] Дж. Эмсли. Элементы. 1993г [19] Ligand view from RCSB PDB site
1. Mikhail Ivanov English translation, description for hydrogen bonds and cation-pi stacking interactions, script for ligands. 2. Egor Kosaretskiy Website design, the description of the active site, the salt bridges and ligands. 3. Denis Azhugim Script and description for hydrophobic core, description of π-π interactions and disulfide bridges.