The times of the hords will come, the Carcharodon is here!
We'll dance out everything Gapon didn't dare to dream about
Compsognathi will arise taking to arms against their foes
They are led into the battle by a Tzar of prehistoric times!
Mare And The Corpse-Eyed Toads, "Molokh"
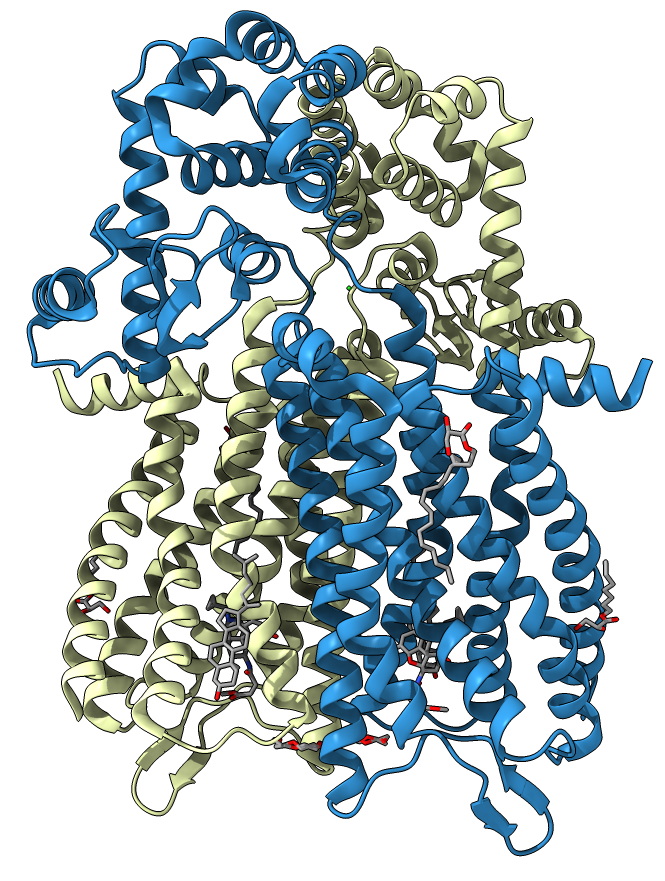
Crystalized protein is a dimeric structure consisting of two similar subunits. Comparison of the μOR-Gi complex to previously determined structures of other GPCRs (G-protein coupled receptors) bound to the stimulatory G protein Gs reveals differences in the interactions between the G protein α subunit and the receptor core: the ligand binds in a deep and open pocket[2].
| Ligand | Structure | Chemical formula | Name by IUPAC | Molecular weight, g/mol | PubChem ID | |
| SO4 | 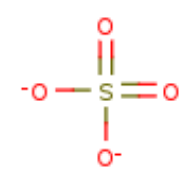 |
SO4 | SULFATE ION | 96.07 | 1117 | |
| Cl |  |
Cl | CHLORIDE ION | 35.45 | 312 | |
| MPG | 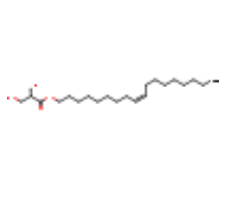 |
C21H40O4 | [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | 356.5 | 17754086 | |
| BF0 | 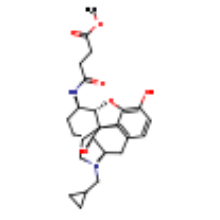 |
C25H32N2O6 | methyl 4-{[(5beta,6alpha)-17-(cyclopropylmethyl)- 3,14-dihydroxy-4,5-epoxymorphinan-6-yl]amino}-4-oxobutanoate |
456.5 | 137348997 | |
| 1PE | 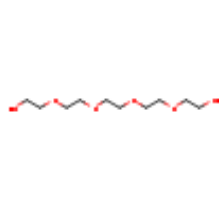 |
C10H22O6 | PENTAETHYLENE GLYCOL | 238.28 | 62551 | |
| CLR | 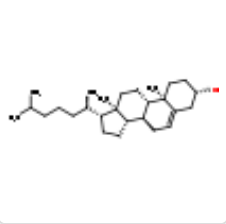 |
C27H46O | CHOLESTEROL | 386.7 | 5997 |
To find covalent sulfur bonds we isolated all the cysteine aminoacids in the protein and manually searched for cysteines forming a bond.
We found one cystine bridge formed by Cys 140 and Cys 217. It can be seen in applet if you choose the «Cystine bridge» script. The bond stabilizes the extracellular domain of the protein while leaving the active site open.
The hydrogen bonds were calculated after restricting all the atoms not involved in the secondary structures like helices and sheets.
In total, the protein has a lot of alpha-helices supported by hydrogen bonds. Beside those hydrogen bonds, there are ones between aminoacid residues not involved in helices and sheets. For example: Thr 1054 and Val 1057, Asn 1020 and Tyr 1024. There are a total of 14 hydrogen bonds not involved in secondary structures. Also there is a bond between Thr 249 of two subunits. These bonds can be seen in the applet when the «Hydrogen bonds» script is chosen. The characteristics of those bonds (Table 2) are supported by those already estimate in reliable sources.[12, 13]
The functional role of these bonds is in stabilizing the dimer structure of the receptor.
| Interacting aminoacids | Atoms participating in a bond | Length, Å | Angle, deg | |
| Thr 249:1 - Thr 249:2 | O - O | 2.6 | 134.5 | |
| Thr 1054 - Val 1057 | O - N | 3.2 | 145 | |
| Asn 1020 - Tyr 1024 | N - O | 2.8 | 142.8 | |
| Asn 1020 - Tyr 1024 | O - N | 2.4 | 111.7 |
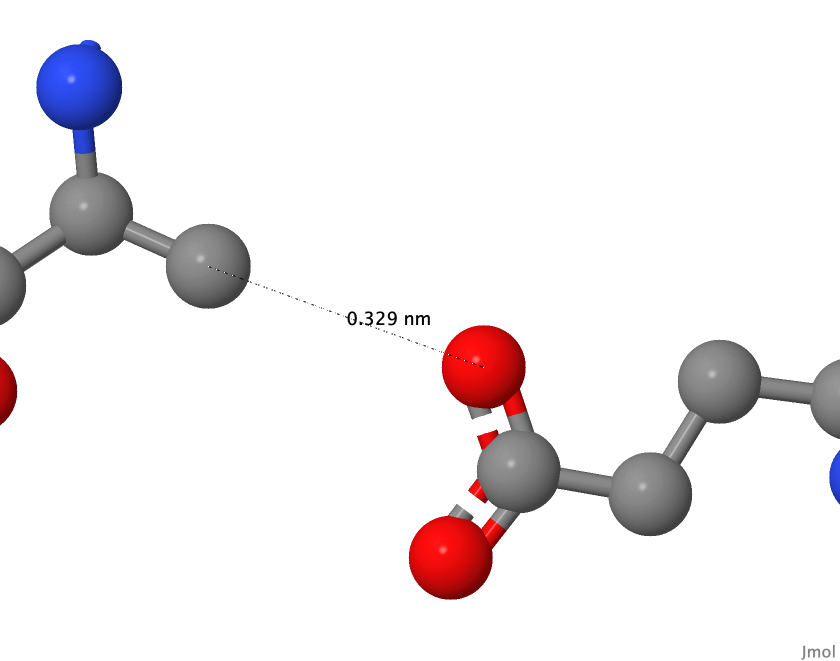
| Residues that form bridge | Distance, Å | |
| 1 | Glu 341 - Lys 344 | 3.5 и 2.9 |
| 2 | Glu 229 - Lys 233 | 3.2 |
| 3 | Glu 1128 - Arg 1125 | 3.5 и 2,8 |
| 4 | Glu 270/1 - Arg 263/2 | <3.3 |
Phe 1153 was chosen to demonstrate the hydrophobic core.
When the script from the application has worked you can see that the distance of 7 Å is the least distance to fully cover the aminoacids with other atoms. On average, the distance between the nuclei of neighboring non-covalently bound atoms in the protein is not more than 6.1 Å, meaning that a water molecule cannot fit into it (distance between the surfaces of two atoms is not more than 2.2 Å, and the diameter of the oxygen atom is 2.8 Å[16]. However there is a hole that is not occupied by anything and that can fit a water molecule in proximity of the sulfur atom of Met 1102.
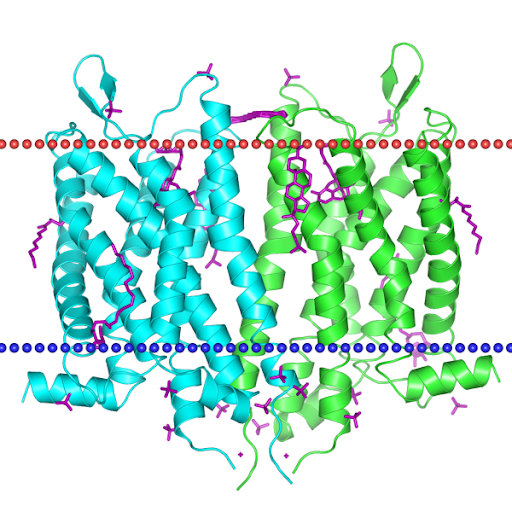
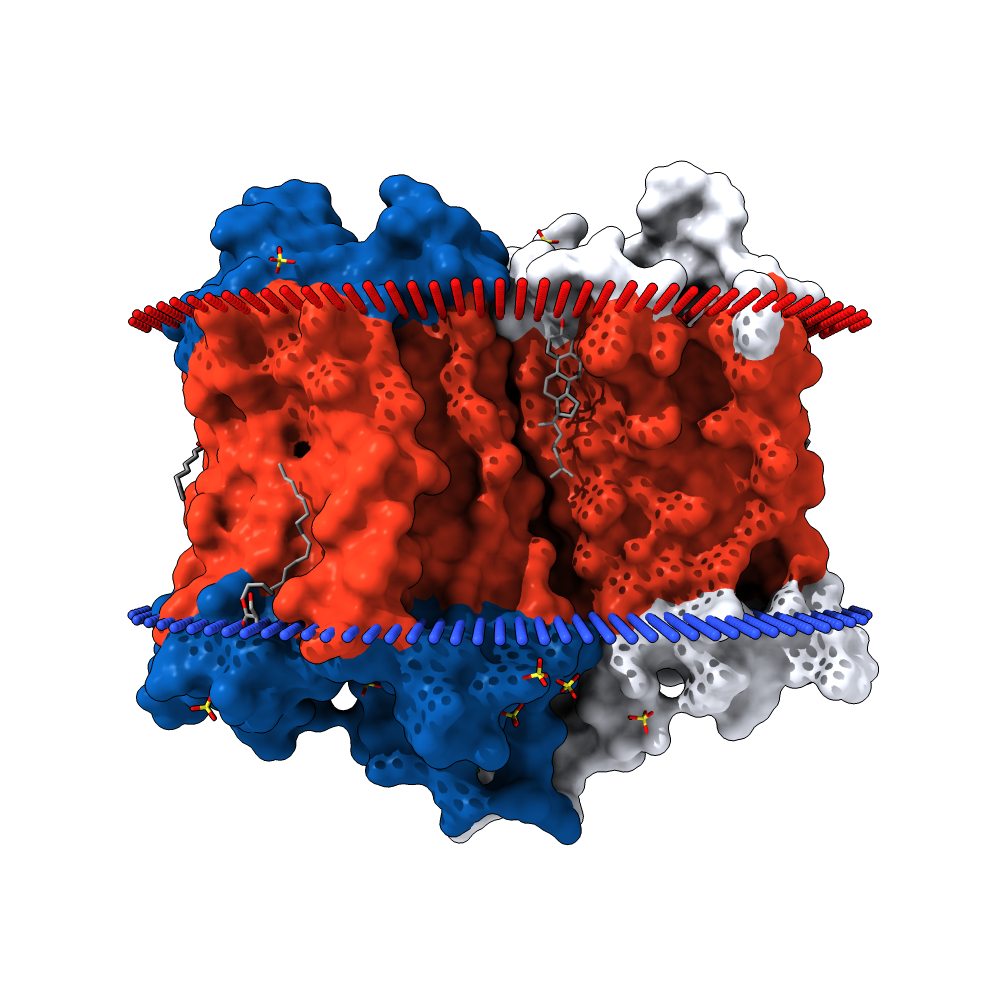
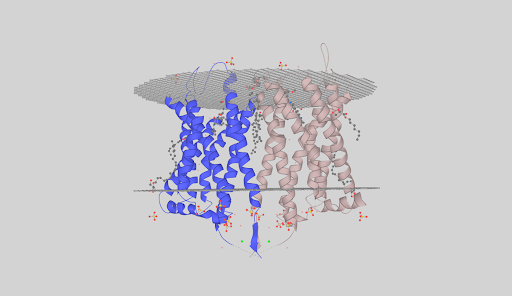
The protein is a GPCR. As stated by the TMHMM prediction (Pic. 4) the protein has 7 transmembrane domains in froms of alpha-helices pointing with the N-end outside the cell. The aminoacids distribution near the membrane is shown in the Table 4. Obviously the protein has inner, outer and transmembrane domains. Inner domains are needed to connect to the G-protein. The part of the protein between the 206 and 384 residue is the largest of all the inner domains and is most likely to be the binding site of the G-protein. Free energy of transmembrane helix association is −65 kcal/mol, which is energetically beneficial — that’s why the protein is neatly packed in the membrane. Visualizations of the protein in the membrane can be seen in the pictures after the TMHMM prediction.
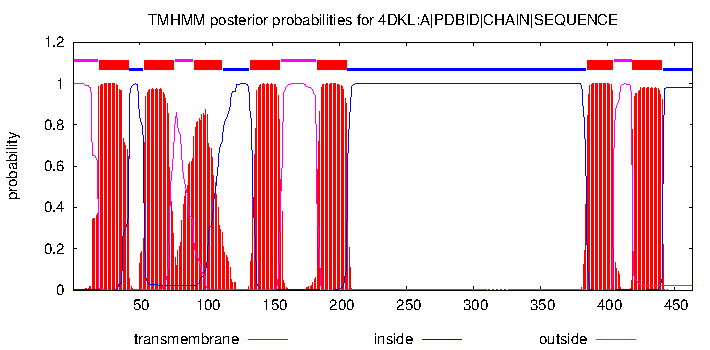
| Extracellular aminoacids |
1- 19 |
77- 90 |
156- 182 |
405- 418 |
|||||||||||
| Transmembrane aminoacids |
20- 42 |
54- 76 |
91- 112 |
135- 155 |
183- 205 |
385- 404 |
419- 441 |
||||||||
| Intracellular aminoacids |
43- 53 |
113- 132 |
206- 384 |
442- 464 |
Dmitry Bosov: ligands application scripts, formulated the priceless contribution of Evgeny Egorov, report translation to the emoji-language
Evgeniy Egorov: most part of the work is done by our Dota 2 esports coach. It is hard to point out any key moments in his actions because any of his advice steers us. For a long time we could not understand how to move on the map. His qualitative analysis was viewing all our solo matches (with random players in the usual game mode) and also our team games. He gave us many tips regarding this aspect, these include:
– arrangement of lines: he placed the roles. He explained to us that carry should go to not to the forest, but to the easy lane.
– explained (especially to the 4th position — Artemiy or Shaker) the features of moving on the map during first moments of the game to supports.
– showed the importance of walking with the team after the lining stage.
Not only he debunked our massive mistakes, but the smaller ones too. He confidently underlined all the mistakes. We were sure that such kind of mistakes were familiar to the Tier 1 players, but when he heard this claim he showed us EvilArthas, the best Dota 2 player.
This is the best player who feels the gaming situation very well. He is able to win a game in solo. Besides a quality game situation analysis he never undrestimates his contribution to the game. He was the man who proved the existence of the hidden pool on his example.
When watching through professional plays (like The International) Arthas (EvilArthas) pointed out that there are 3 important aspects in the game: the first and the most important is heroes pick, the second being the team interactions, and the third being a player’s personal skills (e.g. skill to press BKB in time — as EvilArthas said: Bought BKB, didn’t buy BKB press).
The hero picking stage was mentioned intentionnaly — it was this exact stage that Evgeniy Egorov worked on most of the time. We have many combinations of heroes, thanks to his advice on choosing characters we confidently defend the line.
Most important advice of Coach are:
– Don’t pick Коротышка
– Ban Технари and Коротышка
– icking Рубик as support in 2045 makes you go down (EvilArthas is also sure about that)
– Командир Легиона is the worst hero (Best appearance, worst performance)
– Тень has been losing mid lane for 10 years already
Aside from defining weak heroes Coach also picks out Король Гнева, Луна, Ветрорейнджер.
It is hard to imagine where we would be if Evgeniy Egorov wouldn’t agree to train us. My arguments with the support Vladimir N. sometimes would almost come to fights, but thanks to Evgeniy Egorov the arguments stopped at all. When he first came to us we were on the verge of breakup: constant hidden pool, misunderstandings in the team, bad performance from Призрачный Улан. But he alone managed to set us up for the game, he managed to unlock the potential if every one of us. He wasn’t just a coach — he was a psychologist.
We couldn’t get onto the first qualification matches for Dota 2 Major because of the control work on chemistry, but obviously we were top 1 in the CIS. Besides from chemistry we understood that if we go to Chengdu (China) we would miss the colloquium on the calculus. It’s good we took it into account and chose to stay giving a chance to Na`Vi.
I remember him telling me how to defend the hard lane playing Ветрорейнджер in the right way. I understood that I was making a lot of mistakes and had no idea what I was doing. He showed Artemiy P. how to play Землетряс. I won’t be surprised if after Evgeniy’s guidance Artemiy gives a million dollar Echo Slam (that is a reference to The International when the north american team EG made a combination of Echo Slam + Ice Blast killing the whole enemy’s team when they were in the decisive battle on Roshan.
A reasonable question: how does this all connect to our report of Interaction mapping of the protein?
You know, all that was said above isn’t connected to the report, Evgeniy Egorov is just a good coach. In fact, if it was not for his experience, we would hardly have organized and so harmoniously done this work.
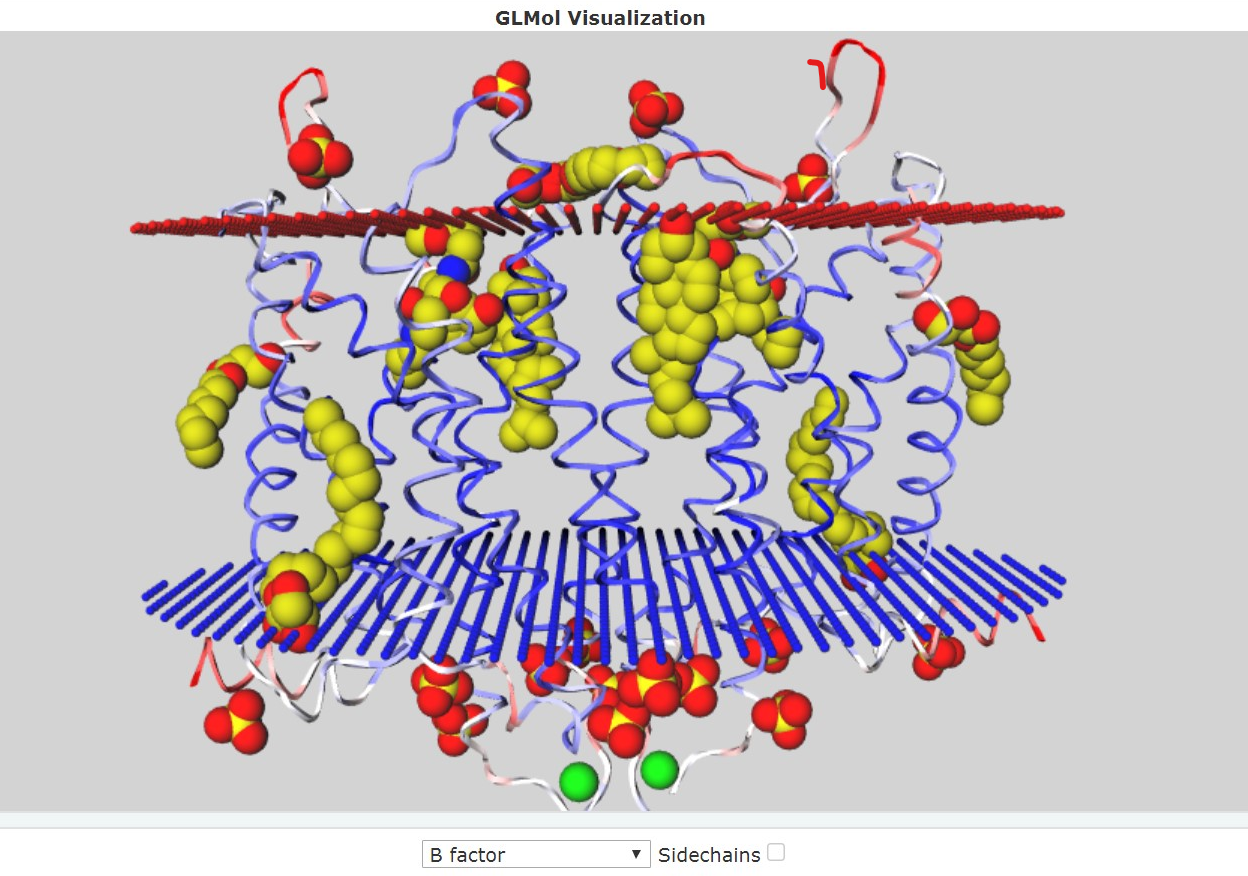
Vladimir Nozdrin: inner protein interactions search, inner protein interactions and hydrophobic core applications scripts, JMol screenshots to accompany the report, creation of the report webpage.
Artemiy Pigidanov: work with TMHMM, visualization of the protein in the membrane by using the OPM, MemProt, GLMol, PDBTM databases, hypotheses considering the functional role of the inner protein interactions found.
Daniil Khlebnikov: writing the report (Ru + En), description of the acquired results, source search, hydrogen bonds search.