|
|
|
- Task 0: New directory was created: /nfs/srv/databases/ngs/nikita/pr14 File: G.fastq
|
|
|
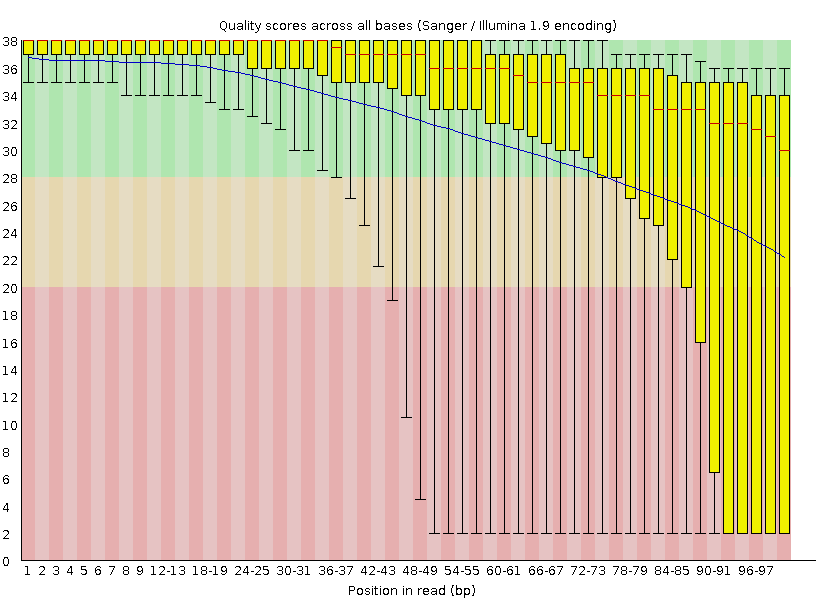
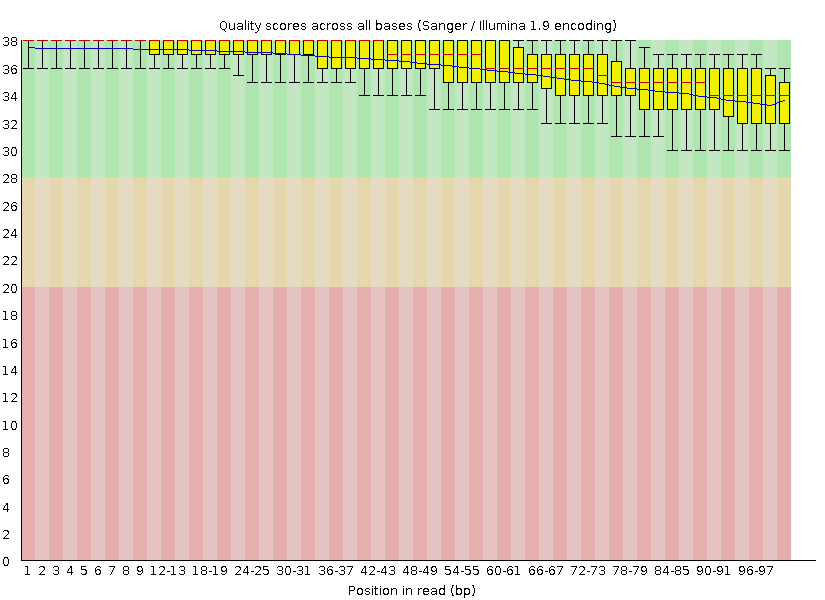
- Task 1: Adapters were coppied from /P/y16/term3/block3/adapters and cat *.* > adapters.fasta was runned to combine them into one file. After that i've used following command to clean up adapters from my file: java -jar /usr/share/java/trimmomatic.jar SE -phred33 G.fastq G_no_adapters.Fastq ILLUMINACLIP:adapters.fasta:2:7:7. Than fastqc G.fastq command was runned. After what i managed to remove the part of reads with the amount quality lower than 28, corresponding command: java -jar /usr/share/java/trimmomatic.jar SE -phred33 G_no_adapters.fastq G_clean.fastq SLIDINGWINDOW:5:28 MINLEN:32.
|
|
|
|
Before (amount of reads - 3869869, 993M)
|
|
|
|
After (amount of surviving reads - 3420075, 797M)
|
|
|
- Task 2: Velveth programm was runned, corresponding command:
- velveth kmers 31 -short -fastq G_clean.fastq
- Velvetg programm was runned, corresponding command:
|
|
| k-mer |
Number of reads |
Maximal contig's lenth and it's coverage |
N50 |
Maximal coverage |
| 31 |
6825516 |
for 606(NODE_316849) - 14.404290, for 590(NODE_49858) - 3.523729, for 589(NODE_29732) - 2.597623 |
28 |
for 41(NODE_2533) - 1478,438965 |
|
- Task 3: Megablast search.
- Megablast search by scaffold's sequences was runned, results:
- Longest scaffold corresponds to Arabidopsis thaliana succinate dehydrogenase 2-2 (SDH2-2), mRNA (Coverage, Identity - both 100%, E-value - 0.00, Accesion - NM_123430.2). Number of alignments - 99
- Second longest scaffold corresponds to Arabidopsis thaliana coiled-coil protein (DUF572), mRNA (Coverage, Identity - both 100%, E-value - 0.00, Accesion - NM_001332684.1). Number of alignments - 34
- Scaffold with maximal coverage corresponds to Arabidopsis thaliana Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family mRNA (Coverage, Identity - both 100%, E-value - 1e-27, Accesion - NM_128266.2). Number of alignments - 11
|
|
|
|