|
|
|
- Proteins-homologues have a common origin.
- Orthologs are proteins that have dispersed as a result of speciation. Thus, orthologic proteins are found in organisms of different species and, as a rule, perform similar functions.
- Paralogues are proteins formed as a result of gene duplication. Consequently, such proteins exist in one organism and evolve almost independently of each other, and therefore can vary greatly in functions.
Task 1:Search for homologues.
- Before the start of work, all bacterial proteomas were downloaded and assembled into a single all.fasta file. The sequence of protein whose homologues have to be found was stored in the file CLPX_ECOLI.fasta.
- Then a database was created from the proteomic file for further homologue searching.
- Command line: makeblastdb -in proteomes.fasta -dbtype prot
- Then the search itself was started using the BLASTP algorithm.
- Command line: blastp -query CLPX_ECOLI.fasta -db proteomes.fasta -evalue 0.001 -out blast.out -outfmt 7 -num_alignments 50
- From the proteins found, I chose the most reliable homologues (the main selection criterion is E-value). The amino acid sequences of these proteins are collected in a file chosen.fasta.
- Next, the alignment of all sequences was constructed.
- Command line: muscle -in chosen.fasta -out align.fasta
Building a tree
- Then a phylogenetic tree was constructed, based on the obtained file, with the Minimum Evolution method
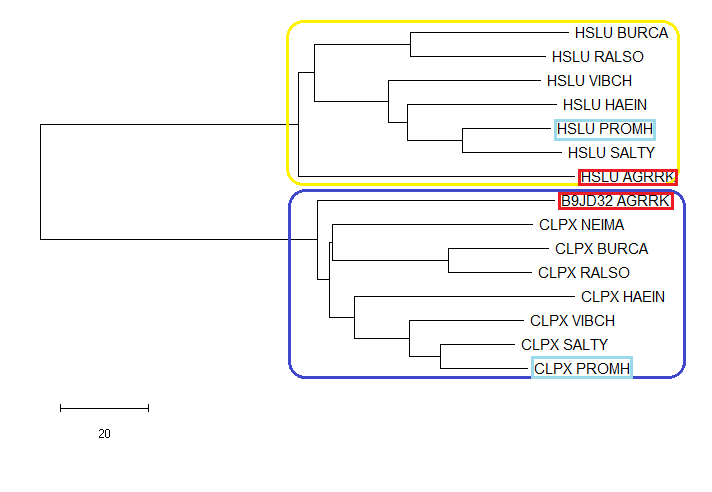 - Area marked blue contains orthologs CLPX and B9JD32. All these proteins are in different organisms, but they perform the same function (ATP-dependent Clp protease ATP-binding subunit ClpX). The yellow color corresponds to the group of orthologous proteins HSLU (ATP-dependent protease ATPase subunit HslU). Pairs of paralogs - proteins, having a common ancestor and located in the body of one species, but functionally separated in the course of evolution, are distinguished by small frames of the same color.
|
|
|
|