|
|
|
- Task 1: To search for motives, it was suggested to find from the bacterium in the Uniprot database 8-10 genes involved in the purine biosynthesis (search for the keyword "Purine biosynthesis"). Below are the data on the proteins found in Shigella dysenteriae SECA (Uniprot-mnemonics - SHIDS).
|
|
| Entry |
Entry name |
Protein names |
Gene names |
Coordinates |
| Q32HB3 |
PURT_SHIDS |
Formate-dependent phosphoribosylglycinamide formyltransferase |
purT SDY_1135 |
1053879...1055057 |
| Q32D15 |
PUR4_SHIDS |
Phosphoribosylformylglycinamidine synthase |
purL SDY_2747 |
complement(2545533...2549420) |
| Q328F6 |
PURA_SHIDS |
Adenylosuccinate synthetase |
purA SDY_4412 |
complement(4127842...4129140) |
| Q32JK7 |
FOLD_SHIDS |
Bifunctional protein FolD |
folD SDY_0281 |
complement(274853..275719) |
| Q32D55 |
GUAA_SHIDS |
GMP synthase [glutamine-hydrolyzing] |
guaA SDY_2703 |
complement(2494237..2495814) |
| Q32AH9 |
PUR9_SHIDS |
Bifunctional purine biosynthesis protein PurH |
purH SDY_3720 |
3450399...3451988 |
| Q32FB3 |
PURR_SHIDS |
HTH-type transcriptional repressor PurR |
purR SDY_1884 |
1718130...1719155 |
| Q32D69 |
PUR5_SHIDS |
Phosphoribosylformylglycinamidine cyclo-ligase |
purM SDY_2688 |
2480836...2481873 |
| Q32D90 |
PUR7_SHIDS |
Phosphoribosylaminoimidazole-succinocarboxamide synthase |
purC SDY_2664 |
complement(2456494...2457207) |
|
- Shigella dysenteriae Sd197, complete genome.
- By coordinates of the coding sequences in the genome, the coordinates of the 100 nucleotides preceding them (from the 5 'end of the start codon) were found. Such nucleotides constitute the Upstream regions of the genes. Further, using the descseq command, upstream regions were gathered into a single file upstream.fasta.
- Then the MEME program was launched, which should find the motives in the sequences. Command line: ememe upstream.fasta -nmotifs 3 -revcomp
- Full result of the MEME program.
- For each motive is given its information content (information content) - a numerical parameter that allows you to evaluate whether the given sequence is a motive or an accidental find. According to rough estimates, the word of length n = I / 2 (where I is the notational content) can occur in the genome once in 4 ^ n pairs of nucleotides.
E-value shows how great the probability of finding with the same or greater weight.
Also shown are LOGO schematic diagrams in which the height of the column is I of this position and shows its contribution to the total I of the entire motif, and the height of the letters is equal to the I column multiplied by the probability of encountering the given letter at that position. Thus, LOGO displays the most plausible sequence of the desired motive.
- Motif 1 (Found between 8 sequences, E-value 2.2e+000, Information Content 16.6 bits)
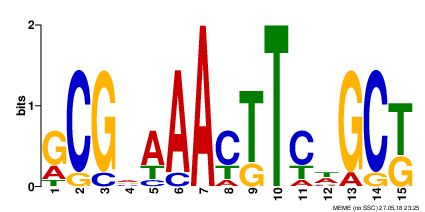
- Motif 2 (Found between 8 sequences, E-value 1.9e+002, Information Content 15.8 bits)
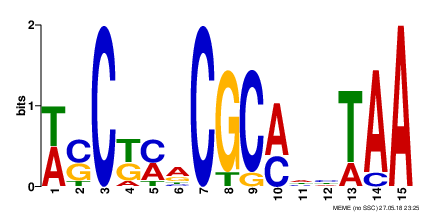
- Motif 3 (Found between 2 sequences, E-value 1.6e+004, Information Content 17.0 bits)
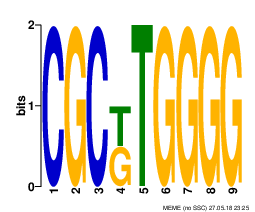
- Each one of the presented motifs can be barely considered as a real one. All of them have high e-value, and not quite high information content
|
|
|
|