|
|
|
- Task 1: For this task P0AD49 ID was choosen (Ribosome-associated inhibitor A). During stationary phase it prevents 70S dimer formation,
probably in order to regulate translation efficiency during
transition between the exponential and the stationary phases
(PubMed:16324148). During environmental stress such as cold shock
or excessive cell density at stationary phase, it stabilizes the 70S
ribosome against dissociation, inhibits translation elongation and
increases translation accuracy (PubMed:11375931, PubMed:15219834).
When normal growth conditions are restored, it is quickly released
from the ribosome
- Data, received from the Blast search are presented in the table below. It can be concluded, that after 3 iterations the result stabilized, the difference between the best finding lower the threshold and the worst one above is more than 20 times, what means that the received selection can be considered as a family of homologous proteins:
|
|
| Iteration number |
The number of findings above the threshold |
Worst finding Id above the threshold |
It's e-value |
Best finding Id below the threshold |
It's e-value |
| 1 |
22 |
Q49VV1.1 |
8e-04 |
P19954.2 |
0.022 |
| 2 |
27 |
O05886.4 |
3e-09 |
Q82IY7.1 |
0.092 |
| 3 |
27 |
O05886.4 |
2e-24 |
Q82IY7.1 |
0.080 |
|
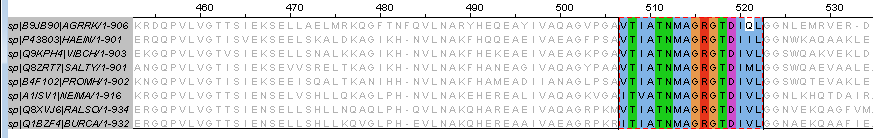
- Task 6: For this task ATPase of Sec-translocon system was used, the search was runned by ID SECA_SALTY. 2 Patterns were found:
- PS01312 (VtIATNMAGRGtDImL, coordinates 500 - 515). Formula: [IV]-x-[IV]-[SA]-T-[NQ]-M-A-G-R-G-x-D-I-x-L
- PS51196 (Coordinates 3 - 619).
- For the futher investigation first pattern was used. New pattern formula: [IV]-T-[IV]-A-T-N-M-A-G-R-G-T-D-I-[VMIQ]-L.
|
|
|
- While comparing with 'right' data it was revealed, that the number of false-positive is almost equal to that of true-positive, while the number of false-negative is quite small.
- Excel file link
|
|
|
|