| FBB MSU site | Main page | About me | Terms |
DNA-protein complexes
Task 1
Exercises 1 and 2
According the task I compared forecast of secondary structure of tRNA (1h3e) made by programe "einverted" (EMBOSS. Minimum score threshold = 5) and by the Zucker's algorithm with forecast mabe by "find_pair" and "analyze" 3DNA package (see pr2). The result you can see in table 1.
* For einverted: Minimum score threshold = 5, "---" means that nothing were find.
Figure 1. tRNA.
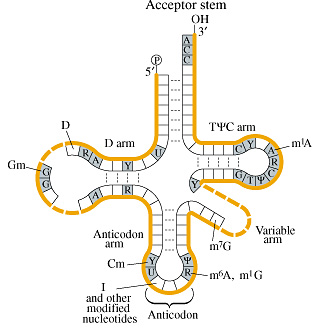
Table 1.
| Part of structure | Forecast by "find_pair" | Forecast by "einverted" | Forecast by Zucker's algorithm |
| Acceptor stem | 5'-1-72 -3' 5'-2-71 -3' 5'-3-70 -3' 5'-4-69 -3' 5'-5-68 -3' 5'-6-67 -3' 5'-7-66 -3' 7 pairs |
7 pairs | 7 pairs |
| D arm | 5'-10-25 -3' 5'-11-24 -3' 5'-12-23 -3' 5'-13-22 -3' 5'-14-21 -3' 5 pairs |
--- | 5 pairs |
| Anticodon arm | 5'-39-31 -3' 5'-40-30 -3' 5'-41-29 -3' 5'-42-28 -3' 5'-43-27 -3' 5'-44-26 -3' 6 pairs |
--- | 6 pairs |
| T arm | 5'-49-65 -3' 5'-50-64 -3' 5'-51-63 -3' 5'-52-62 -3' 5'-53-61 -3' 5 pairs |
--- | 3 pairs of 5 |
| Number of canonical nucleotide pairs | 24 | 7 |
* There are another 3 complementary pairs in variable arm forecasted by Zucker's algorithm. (see fig. 2)
Figure 2. Forecast of secondary structure of tRNA by the Zucker's algorithm.
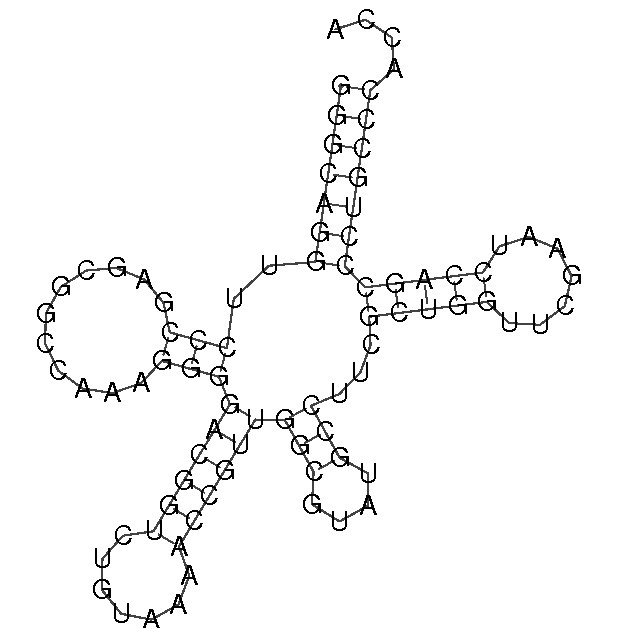
Task 2
Exercise 1
According the task I defined three sets of atoms in my DNA-protein complex (1PP7):
- Set1 - the oxygen atoms of 2'-desoxyribose;
- Set2 - the oxygen atoms in phosphates;
- Set3 - the nitrogen atoms in nitrogen bases.
|
|
Press "Start script" and then "Resume", please. |
Exercise 2
Here oxygen and nitrogen atoms are called polar, and carbon, phosphorus and sulfur atoms are called nonpolar. So I define situation, in which the distance between the polar atom and a polar protein atom DNA less than 0.35 nm, as a polar contact. Similarly, as a nonpolar contact, we will consider a pair of nonpolar atoms at a distance less than 0.45 nm.
The list of contacts you can see in table 2. According pr2 I defined atoms of bases are facing minor or major groove.
To see the visualisation of contacts press "Start script 2" (the text of the script you can see here: script 2. Polar atoms of DNA colored green, polar atoms of protein colored blue, nonpolar atoms of DNA colored purple, nonpolar atoms of protein colored orange.
Table 2.
| There is the contact between protein's atom and ... | Polar | Nonpolar | Sum |
| ... atoms of 2'-desoxyribose | 5 contacts:
| 23 contacts:
| 28 contacts |
| ... atoms of phosphate | 9 contacts:
| 14 contacts:
| 23 contacts |
| ... atoms of atoms in nitrogen bases in major groove | 2 contacts:
| 7 contacts:
| 9 contacts |
| ... atoms of atoms in nitrogen bases in minor groove | 3 contacts:
| 1 contacts:
| 4 contacts |
Exercise 3
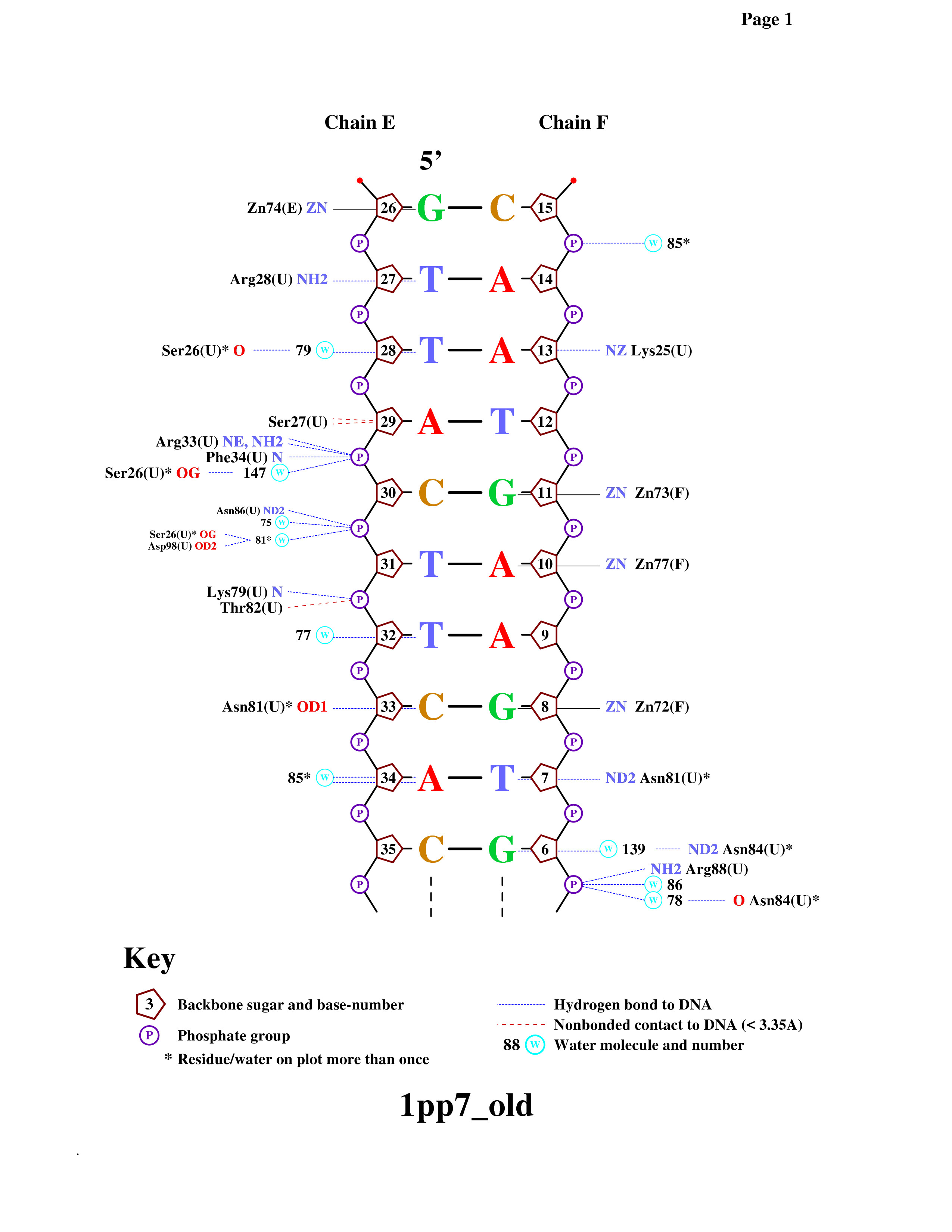
To get a diagram of the DNA-protein contacts I used the program nucplot.
Syntax:
remediator --old ''1pp7.pdb'' > ''1pp7_old.pdb''
nucplot 1pp7_old.pdb
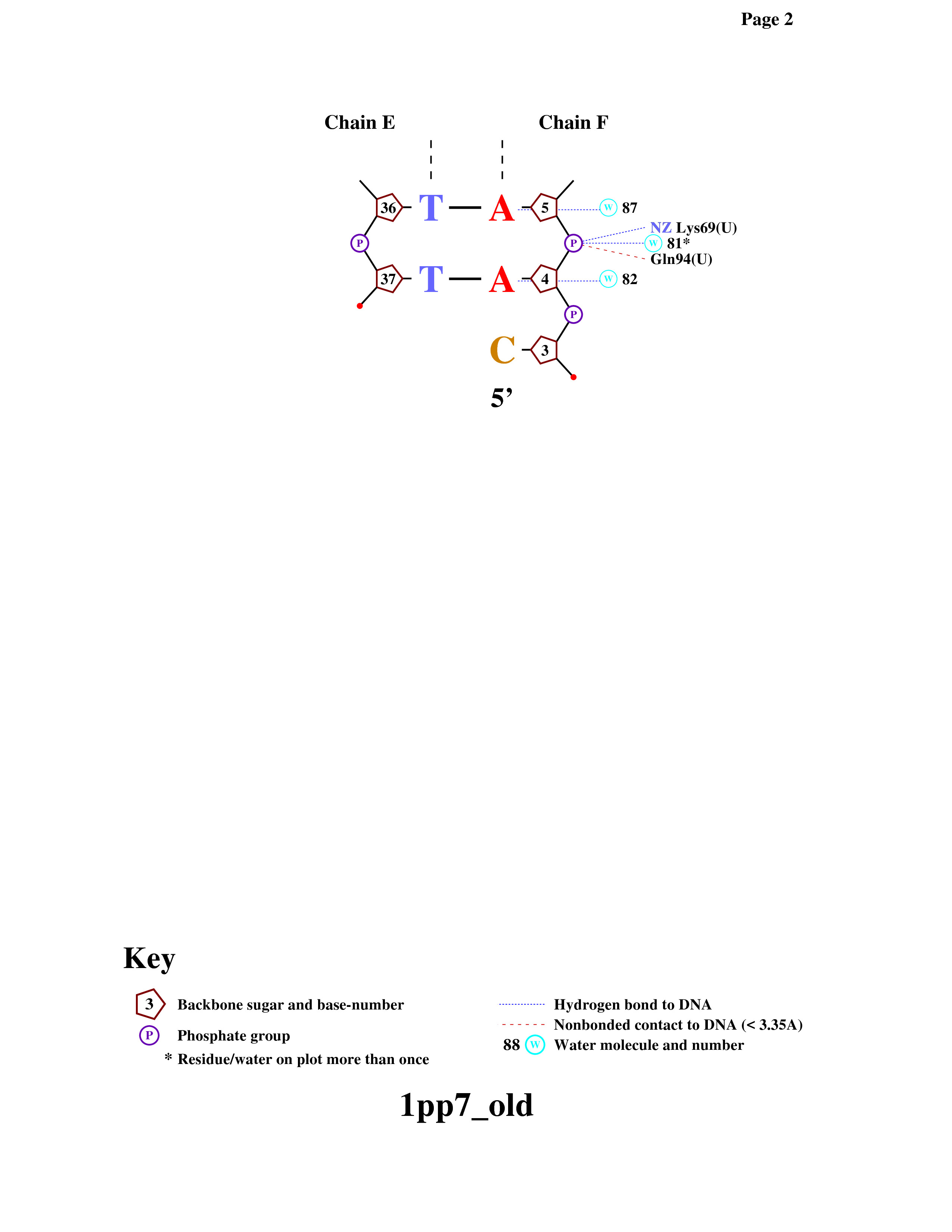
Then I regained the diagram from .ps-file. The result you can see on the figure 3 or here: contacts.pdf.
Figure 3. DNA-protein contacts diagram.
 |  |
Exercise 4
4.1) The amino acid residue which forms the greatest number of hydrogen bonds is Asn81(U): it conects with 33 C and 7 T. The amino acid residue which forms the greatest number of nonbonded contact is Ser27: it conects with 29 A by two bonds. (see fig. 3 and fig. 4)
Figure 4. The amino acid residue which forms the greatest number of contacts.
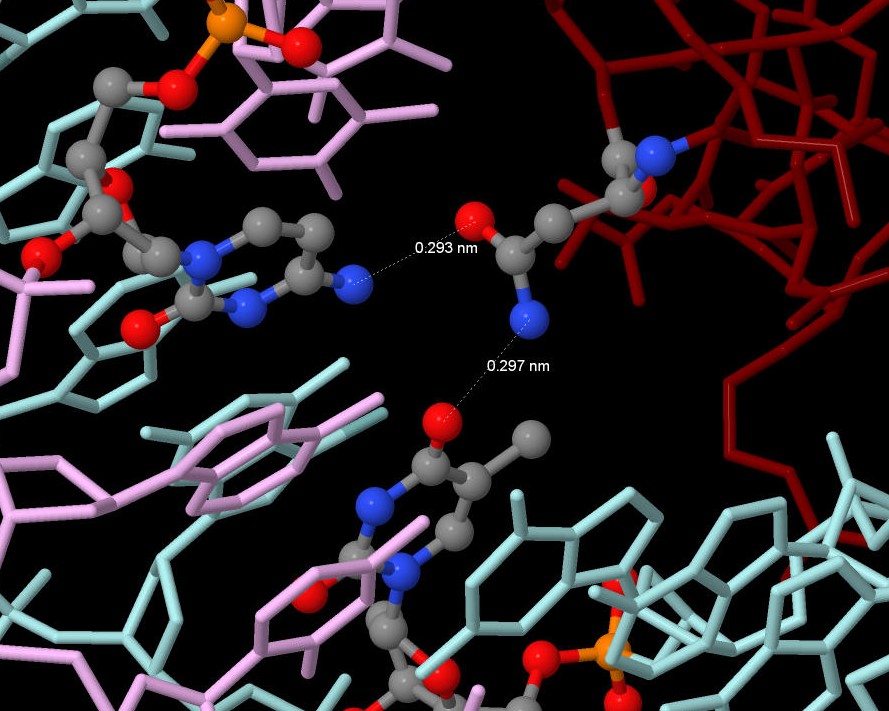 | 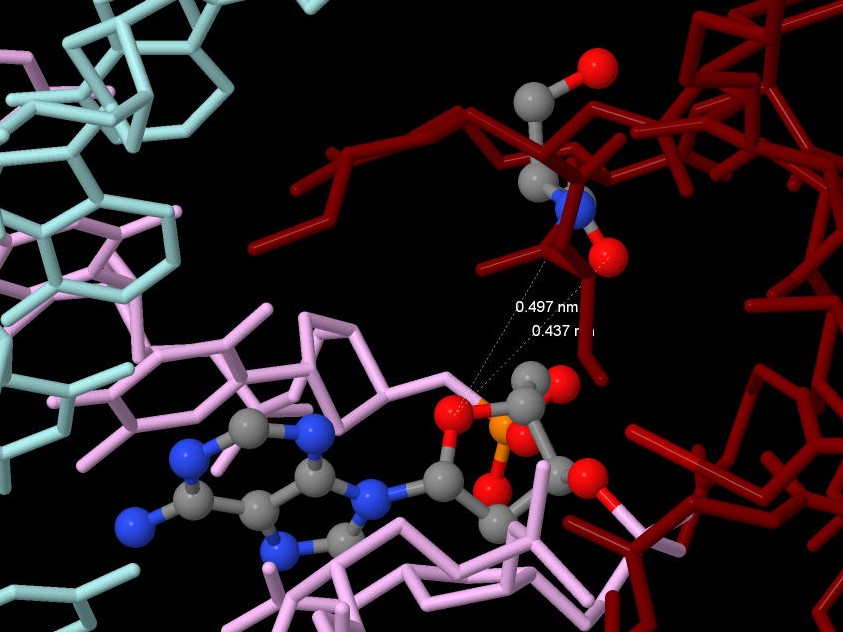 |
4.2) I think the protein recognizes the region 30-CTT-32 (chain E) of DNA by aminoacids: Arg33(U), Phe34(U), Asn86(U), Lys79(U), Thr82(U). I consider so, because every phosphate of these nucleotides is linked with amino acids: 33 C - Arg33(U) and Phe34(U), 31 T - Asn86(U), 32 T - Lys79(U) and Thr82(U) (see fig. 3 and fig. 5).
Figure 5. The aminoacid residues which recognize DNA.
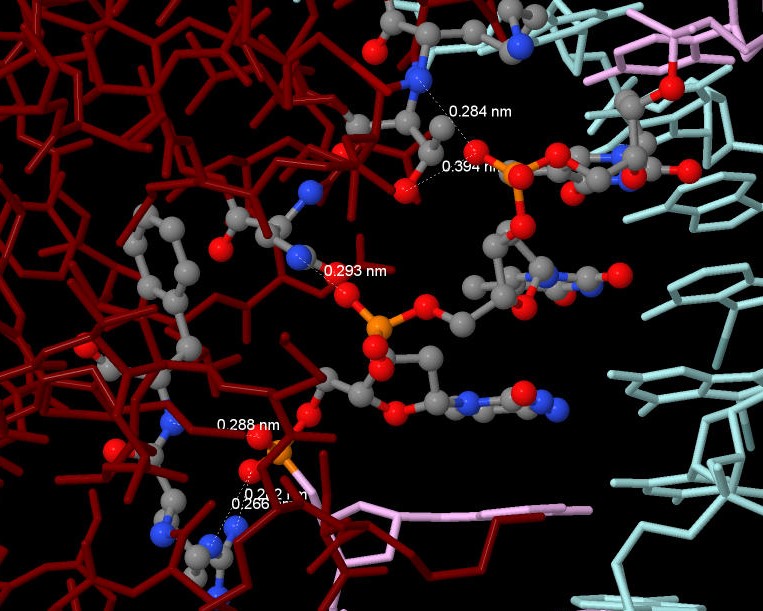
| Term 3 |
| ← Pr 2 | Block 2 → |
© Darya Potanina, 2017