| FBB MSU site | Main page | About me | Terms |
Structure of DNA and RNA
Task 1
According the task we have to create three files with description of 3D structure of A-, B- and Z-DNA forms. The sequence which we used was GATC repeat four times. But for Z-DNA we used GC repeat 10 times, because there isn't any information about A and G in Z_DNA. We used the program "fiber" 3DNA package. There are links to files with completed assignments: A-DNA, B-DNA, Z-DNA. You can see the visualisation of these DNA forms on fig 1.
Figure 1.
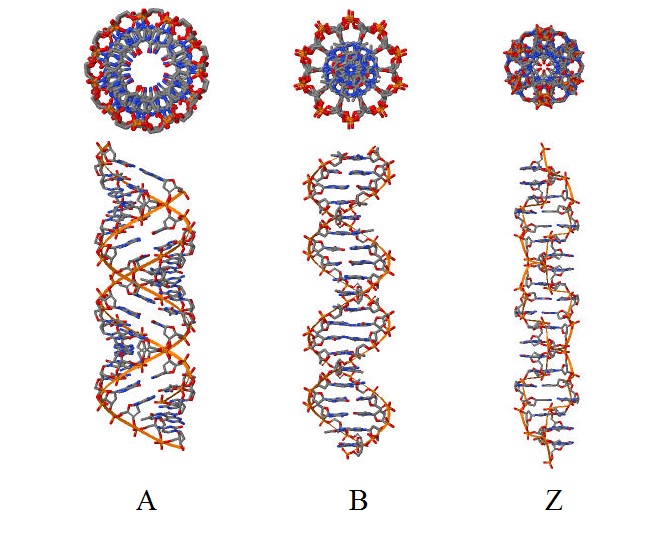
Task 2
Exercise 1
For visualisation I opened the file of B-DNA in JMol and chose a thymine. Then I studied which atoms of this thymine are facing minor groove and which facing major groove. List of these atoms you can see below and on fig 2 (made with MarvinSketch).
Figure 2. |
|
Exercise 2
We studied parametres of minor and major grooves in differend DNA forms with JMol. The result you can see in the table 1.Table 1.
| A-DNA form | B-DNA form | Z-DNA form | |
| Type of helix | right | right | left |
| Pitch of helix | 28,03 Å | 33,75 Å | 43,50 Å |
| Number of bases included in one turn of the DNA helix | 11 | 10 | 12 |
| Width of minor groove | 7,98 Å (between [DG]13:A.P #247 and [DG]21:B.P #411) |
11,69 Å (between [DC]12:A.P #228 and [DG]33:B.P #657) |
9.87 Å (between [DG]11:A.P #206 and [DC]34:B.P #679) |
| Width of major groove | 16,81 Å (between [DC]32:B.P #638 and [DC]12:A.P #228) |
17,21 Å (between [DG]13:A.P #493 and [DG]13:A.P #247) |
18,30 Å (between [DC]10:A.P #187 and [DC]28:B.P #556) |
* The width of grooves was measured as a local minimum of distance between P in different complementary DNA strands in the groove.
Task 3
Exercise 1
I studied 3D srtucture of tRNA 1h3e, DNA 1pp7 and A-, B-, Z-DNA which was created earlier (see Task 1). To determine torsion angles, hudrogen bonds and other parametres of their structure I used the programs "find_pair" and "analyze" 3DNA package. Then I examined files XXXX.out and calculated average values of the angles and found the most deviating values using Excel(see table 2). The excel file with completed ex. 1 you can download here.
Figure 3. Torsion angles of nucleotide.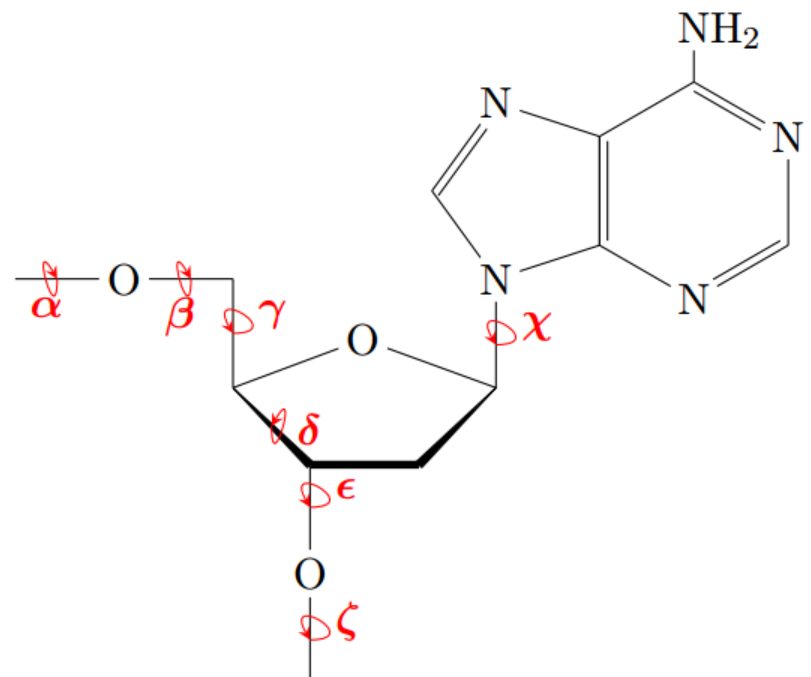 |
Table 2.
|
- The values of the torsion angles for tRNA similar to values for A-DNA.
- There are some nucleitides with the most deviating values of the torsion angles (see table 3). Them were found for the maximum number of the maximum deviation from average values.
Table 3.
α (alpha) β (beta) γ (gamma) δ (delta) ε (epsilon) ζ (zeta) χ (chi) tRNA Strand I
21 U-48.90 174.60 41.30 81.00 -146.90 -67.60 -163.40 tRNA Strand II
7C-57.60 -176.40 47.40 81.90 -147.20 -74.80 -159.60 DNA Strand I
9 A-44.00 152.00 41.80 142.30 -176.30 -62.20 -78.20 DNA Strand II
9 T-48.60 -177.90 37.80 130.30 -158.00 -106.30 -113.10
Exercise 2
There is list of hydrogen bonds in tRNA (see table 4).
Table 4.
| Strand I | Strand II | Helix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Accorditg to table 5 I decided that there are stems in tRNA structure builded by:
- Nucleotides 1-7 associate with 66-72
- Nucleotides 49-54 associate with 65-58
- Nucleotides 35-44 associate with 33-26
- Nucleotides 10-14 associate with 25-21
There are some noncanonic pair: 54 5MU(T) and 58 1MA(A), 55 PSU(P) and 18 G, 37 A and 32 C, 44 U and 26 G, 13 G and 22 A 22, 14 A and 21 A, 47 G and 47 U.
Additional hydrogen bonds stabilizing the tertiary structure of RNA: 55 PSU(P) and 18 G, 15 G and 48 C, 19 G and 56 C, 47 C and 47 E G, 47A G and 47B U.
And there is something very strange (see last few rows in the table 4): 20 A and 47H U, 45 G and 47G C, 46 G and 47F C.
| Term 3 |
| ← Pr 1 | Pr 3 → |
© Darya Potanina, 2017