| FBB MSU site | Main page | About me | Terms |
Signals, motifs, PWM
Preliminary work
According the task 9 proteins engaged in purine biosynthesis were chosen:
keyword:"Purine biosynthesis [KW-0658]"
organism:"Yersinia pestis"
There are 45 reviewed and there are 5 popular organisms. Among them I chose Yersinia pestis bv. Antiqua (strain Antiqua) which have 9 reviewed proteins. Mnemonic: YERPA. EMBL: CP000308 Genomic DNA.
The list of reviewed proteins of Yersinia pestis bv. Antiqua you can see in the folloing table.
| Entry | Entry name | Protein names | Gene names | Coordinates | + 100 nt upstream |
| Q1C5E7 | PUR4_YERPA | Phosphoribosylformylglycinamidine synthase, FGAM synthase, FGAMS, EC 6.3.5.3 | purL YPA_2360 | complement(2646172..2650035) | complement(2650036..2650136) |
| Q1C8V8 | PURT_YERPA | Formate-dependent phosphoribosylglycinamide formyltransferase | purT YPA_1147 | complement(1300219..1301400) | complement(1301401..1301501) |
| Q1C105 | PURA_YERPA | Formate-dependent phosphoribosylglycinamide formyltransferase | purA YPA_3906 | complement(4398153..4399451) | complement(4399452..4399552) |
| Q1C4U4 | FOLD_YERPA | Bifunctional protein FolD | folD YPA_2565 | 2858239..2859105 | 2858138..2858238 |
| Q1C5J6 | GUAA_YERPA | GMP synthase [glutamine-hydrolyzing], EC 6.3.5.2 | guaA YPA_2311 | complement(2587010..2588587) | complement(2588588..2588688) |
| Q1C1V9 | PUR9_YERPA | GMP synthase [glutamine-hydrolyzing], EC 6.3.5.2 | purH YPA_3601 | 4038210..4039799 | 4038109..4038209 |
| Q1C1V9 | PURR_YERPA | HTH-type transcriptional repressor PurR | purR YPA_1732 | 1942437..1943462 | 1942336..1942436 |
| Q1C5P2 | PUR5_YERPA | Phosphoribosylformylglycinamidine cyclo-ligase, EC 6.3.3.1 | purM YPA_2265 | 2536453..2537496 | 2536352..2536452 |
| Q1C5Q7 | PUR7_YERPA | Phosphoribosylaminoimidazole-succinocarboxamide synthase, EC 6.3.2.6 | purC YPA_2250 | complement(2523727..2524440) | complement(2524441..2524541) |
Sequences of upstream regions was cut from FASTA file with genome and saved in 100upstream.fasta. (Syntax: to cut sequenses: 'descseq CP000308.fasta:[xxxx:xxxx:r?] -name **** -description YPA_**** -outseq up*.fasta', to join them: 'seqret up*.fasta 100upstream.fasta)
To find motifs in upstream regions I used 'ememe' installed on kodomo. Syntax: 'ememe -nmotifs 3 -revcomp 100upstream.fasta' (-nmotifs 3 = find 3 motifs, -revcomp = use both forward and reverse sequences). Output files here and the main one - meme.html.
Analysis of motifs
Width - width, sites - number of occurrences in the training set (number of sequences where motif was found), I - information content, llr - log likelihood ratio, E-value - you understand, it is reliability of our findings.
MOTIF 1
|
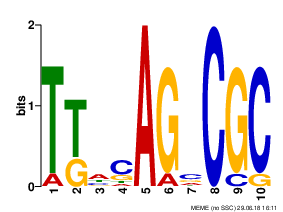 |
MOTIF 2
|
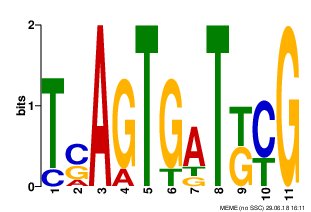 |
MOTIF 3
|
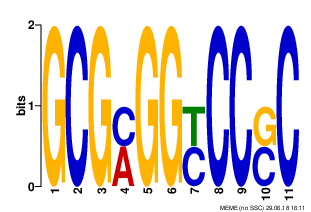 |
Summary:
Motives 1-3 have E-value > 0.001 and low information content. Motif 1 only was found in all 9 sequenses. I think these motives are not reliable.
| Term 4 |
| ← Block 1 | → Pr 6 |
© Darya Potanina, 2017