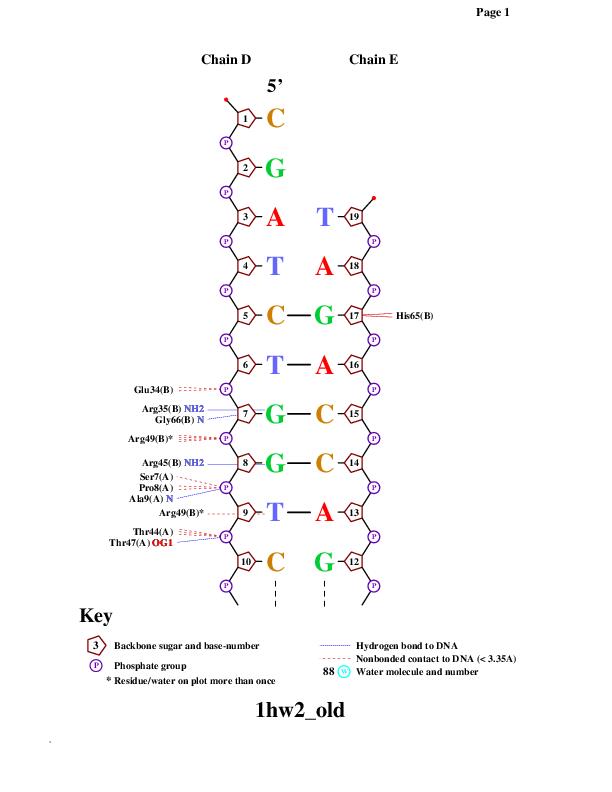
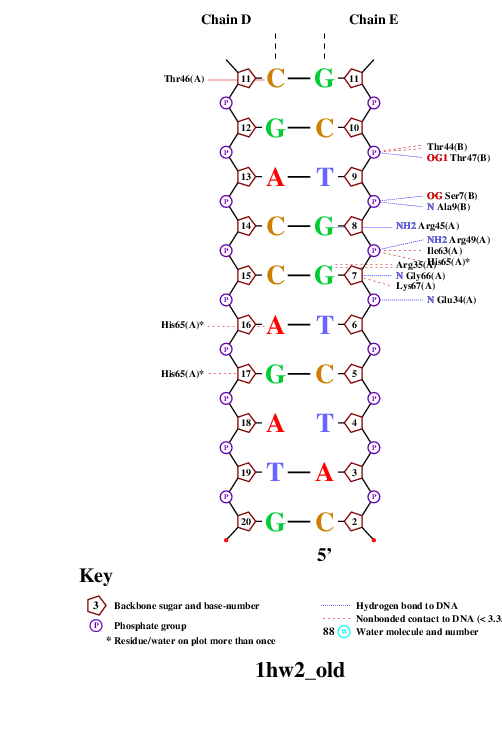
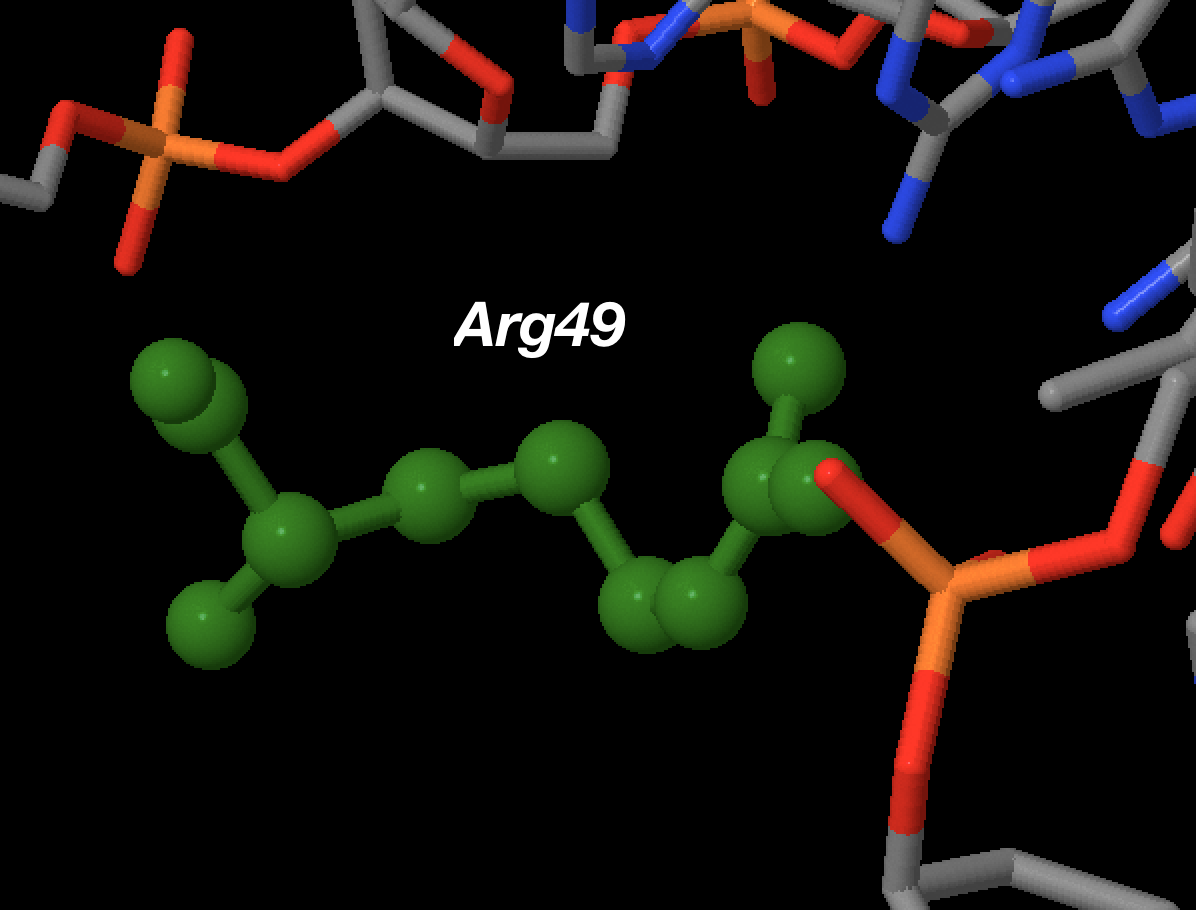
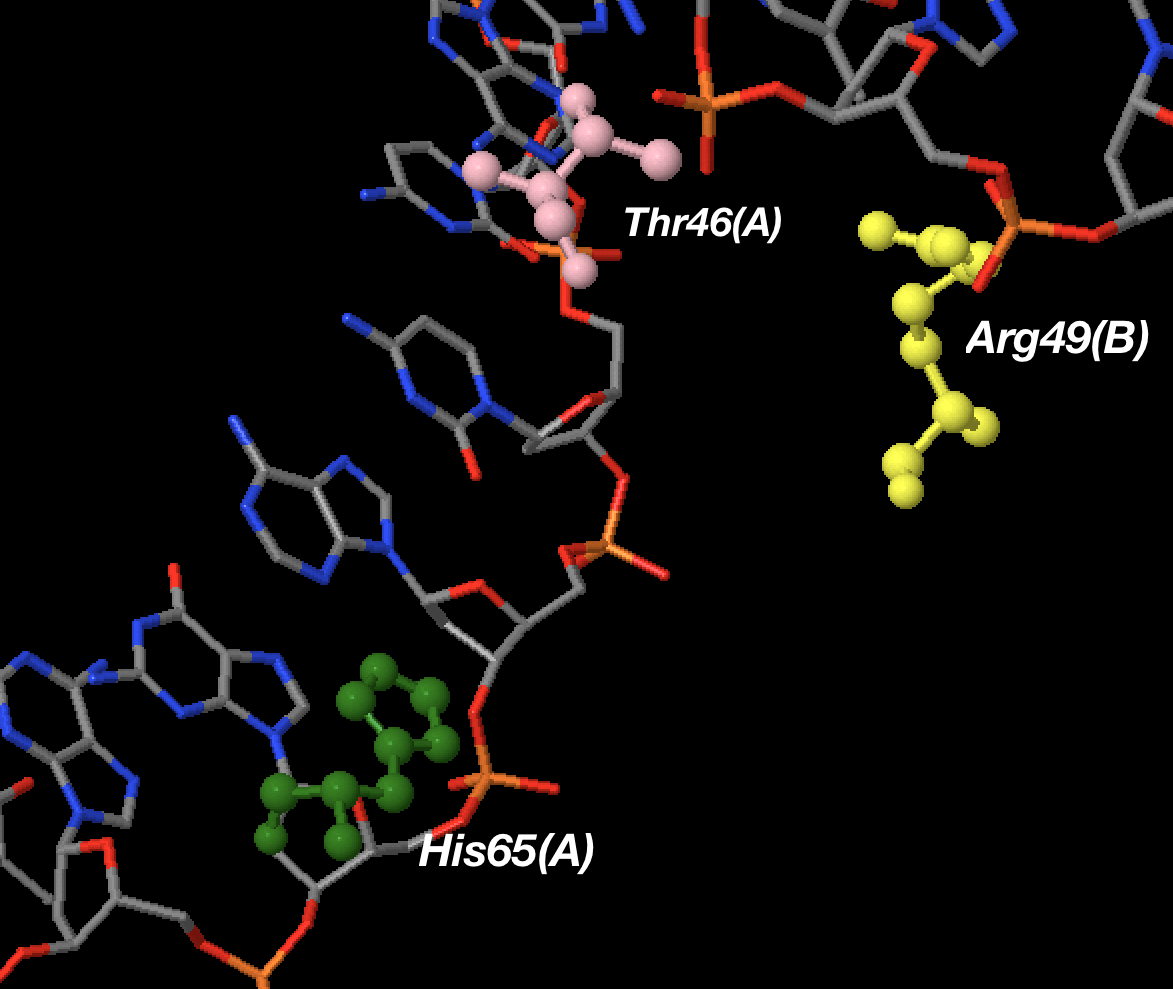
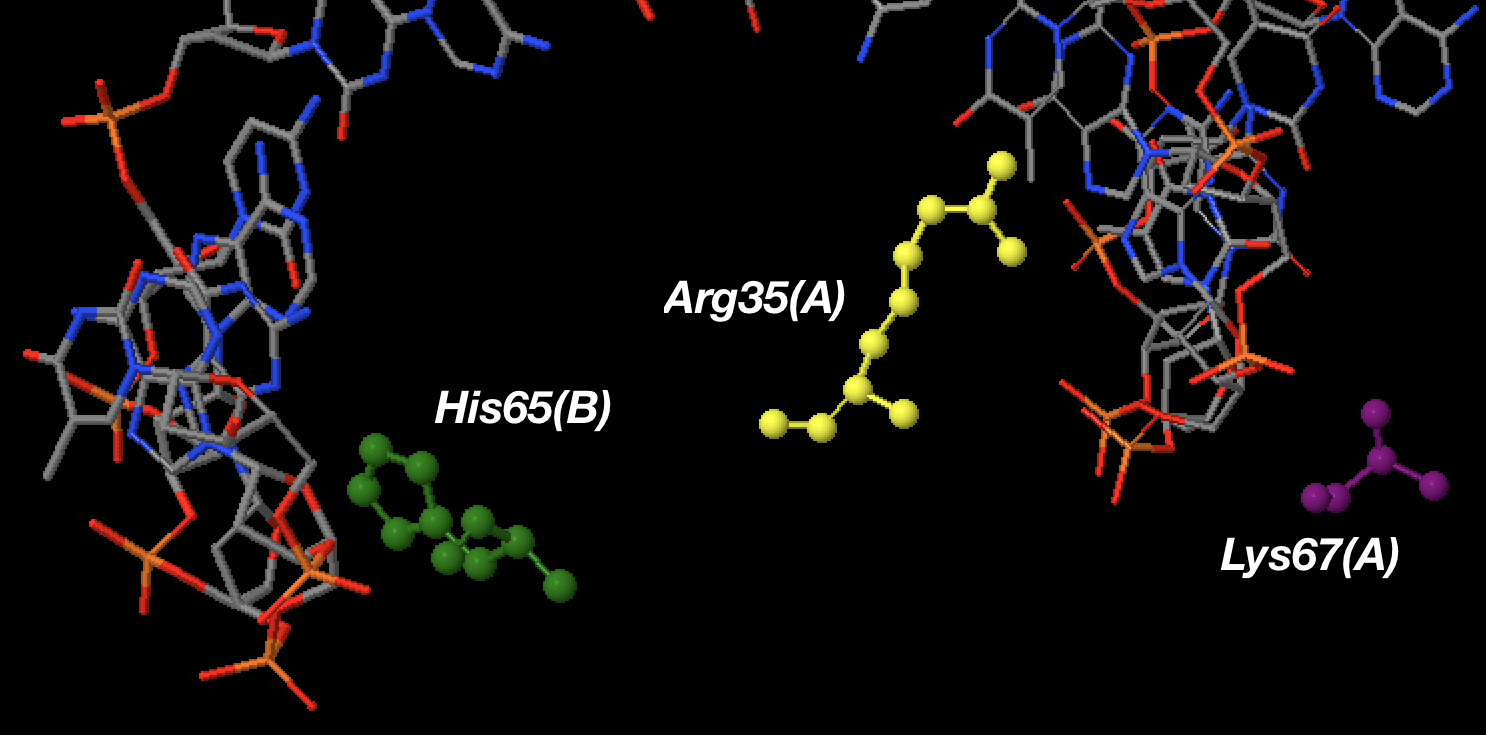
Einverted results
: Score 72: 24/24 (100%) matches, 23 gaps
1 gccgaggta-gc----tcagtt----g-gtagagca-tgcgac---t 33
||||||| || || | | | || || ||||| |
72 cggctcc--gcgccttag-c--ttggcgc--ct-gtgacgct-aaaa 35
Zuker algorithm results
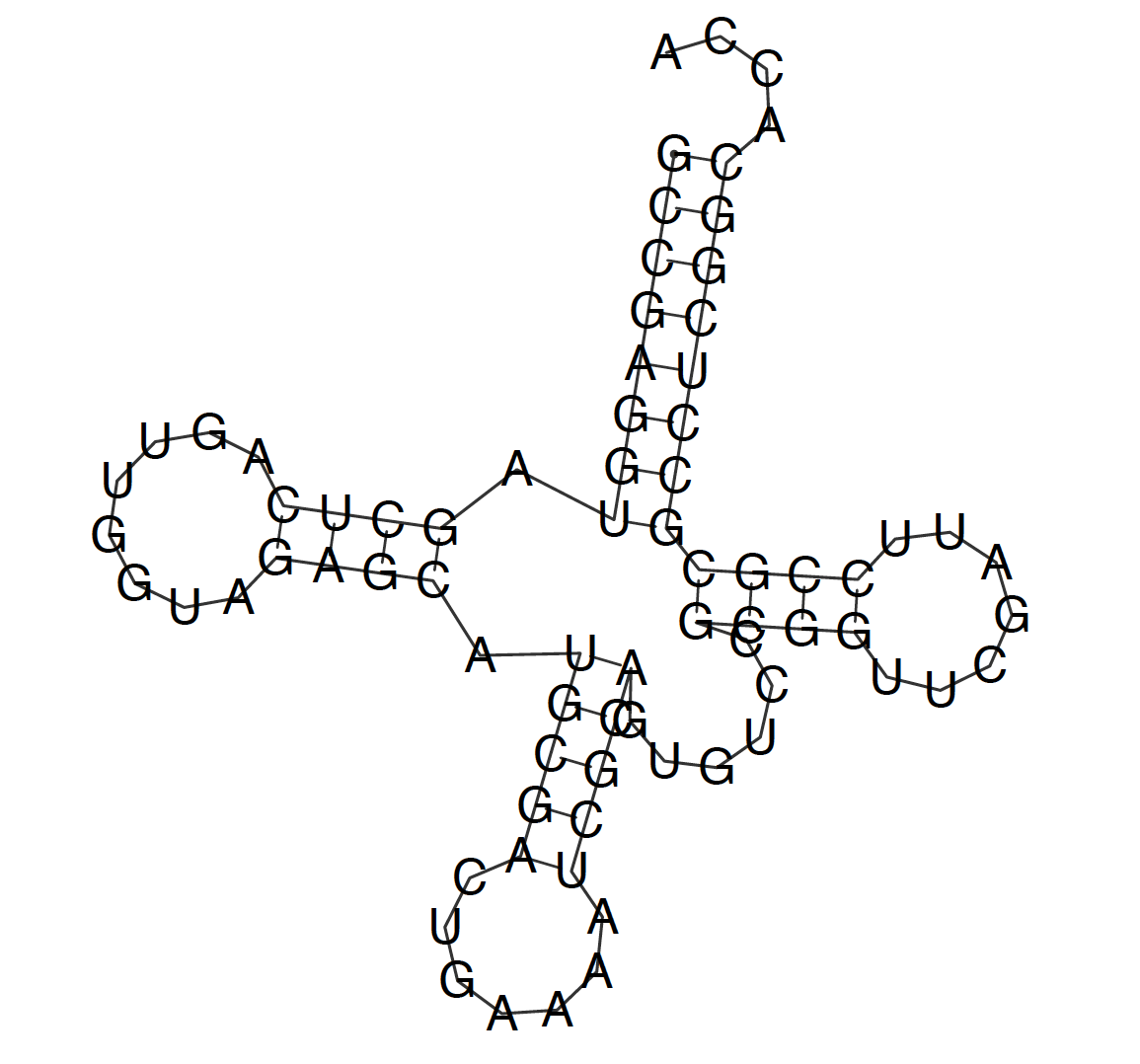
Figure 1. Structure predicted by Zuker algorithm