|
Home page Term 1 Term 2 Term 3 About me Faculty website |
DNA-Protein ComplexesInverted repeatsThis section contains an comparison of different programs (and hence algorithms) predicting locations of tRNA stem helices. These helices are formed in inverted repeat structures, and the calculations the programs perform are based on this fact.The three programs analyzed here are find_pair (3DNA), einverted (EMBOSS) and RNAfold (Viena RNA Package). The latter employs Zuker's algorithm in its calculations.
RNAfold predicted all the elements correctly the first time it was launched. The following image represents this first prediction: 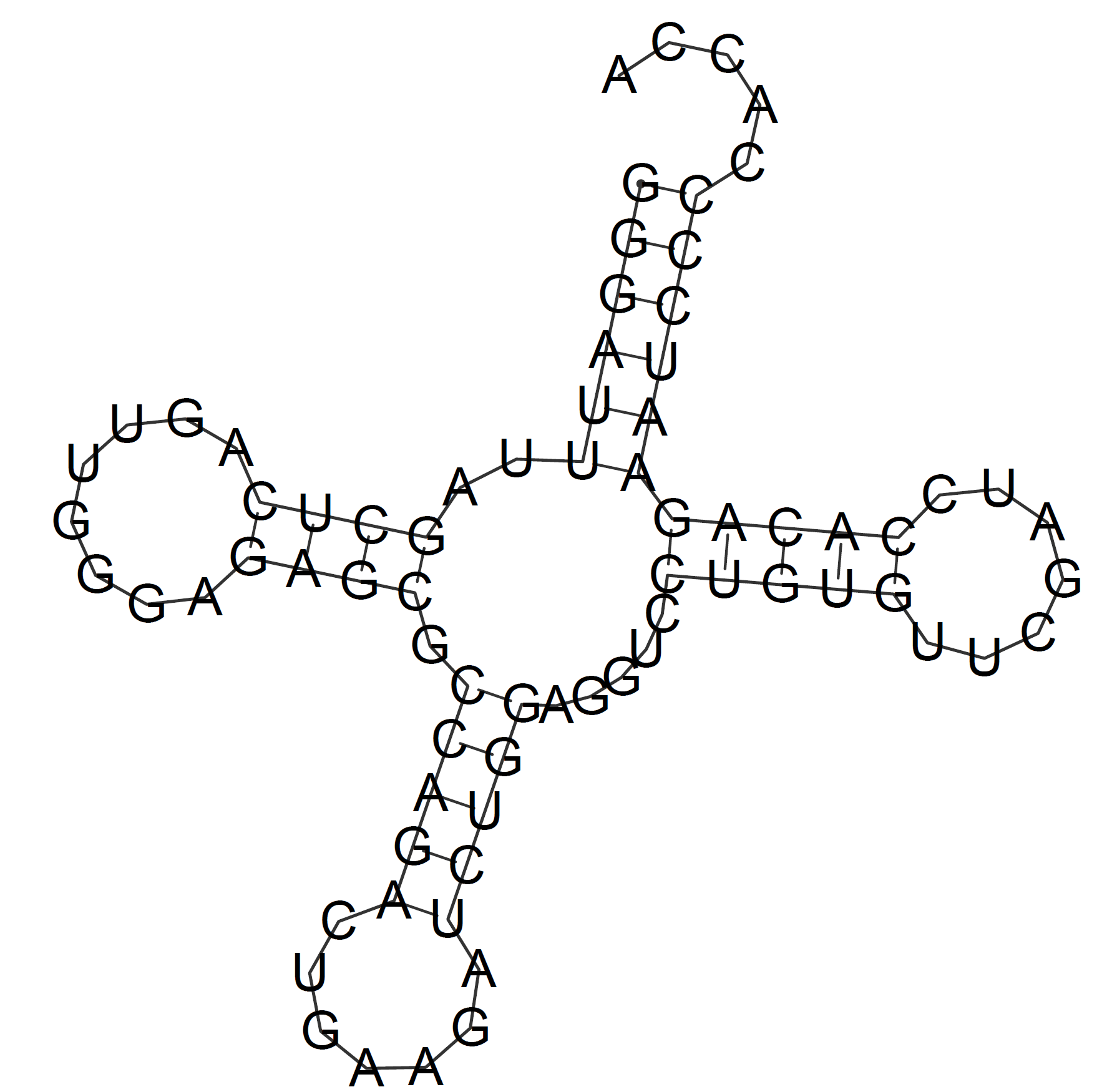
DNA-Protein complex visualized in JMolFor this task the complex with PDB ID "4Z1X" was visualized. Set1 contains all the oxygens pertaining to 2'-deoxyribose, set2 is composed of all the oxygens in phosphate groups, and all the nucleobase nitrogen atoms comprise set3.
DNA-Protein contacts count and analysisThe following table displays the numbers of contacts between different parts of the DNA and protein in the given structure (4Z1X). The numbers were obtained with the aid of a JMol script (there is a link to it below the table).
JMol script The numbers that are added together in the table represent DNA-protein contact counts pertaining to the two DNA helices. In the "Total" column they represent the polar and non-polar contacts added together. The numbers in the brackets (at the end of each table cell) represent the numbers of contacts that I detected using incorrect contact definitions: I used 3.0 Å distance for each type of the contacts. The numbers before the brackets represent contact counts with correct definitions, but in some cases they may be approximate, e.g. 2x(40-50) means that there are from 40 to 50 contacts for each of the DNA helices. The protein has beta-sheet motifs taylored to specifically bind the major groove of the DNA helix. The protein is a dimer, but each monomer forms different number of contacts with the DNA in this experimental structure (there is a dimer bound to each DNA helix in the structure). Also, alpha-helices are involved in hydrophilic contact formation, but to a much lesser extent. As it can be seen from the table, the DNA-protein interface in this structure is hydrophilic, and, consequently, all the contacts are between the "polar" atoms. NucplotThis is a .pdf file that was given out by the nucplot program. There are plenty of amino acids which formed two contacts with the DNA and zero amino acids with three or more contacts. Although it is rather pointless to try to select the one amino acid which is the most important for the DNA sequence recognition (sequence recognition happens because of a multitude of contacts forming at the same time), I think that Lys42(B) can be a candidate for this title. It forms two hydrogen bonds and each one with a different nucleobase, hence recognizing the sequence. Then it draws Glu70(B) towards itself, stabilizing the beta-sheet, which, in its turn, recognizes adjacent parts of the DNA sequence.Below one can see two pictures of the lysine in question: 1) 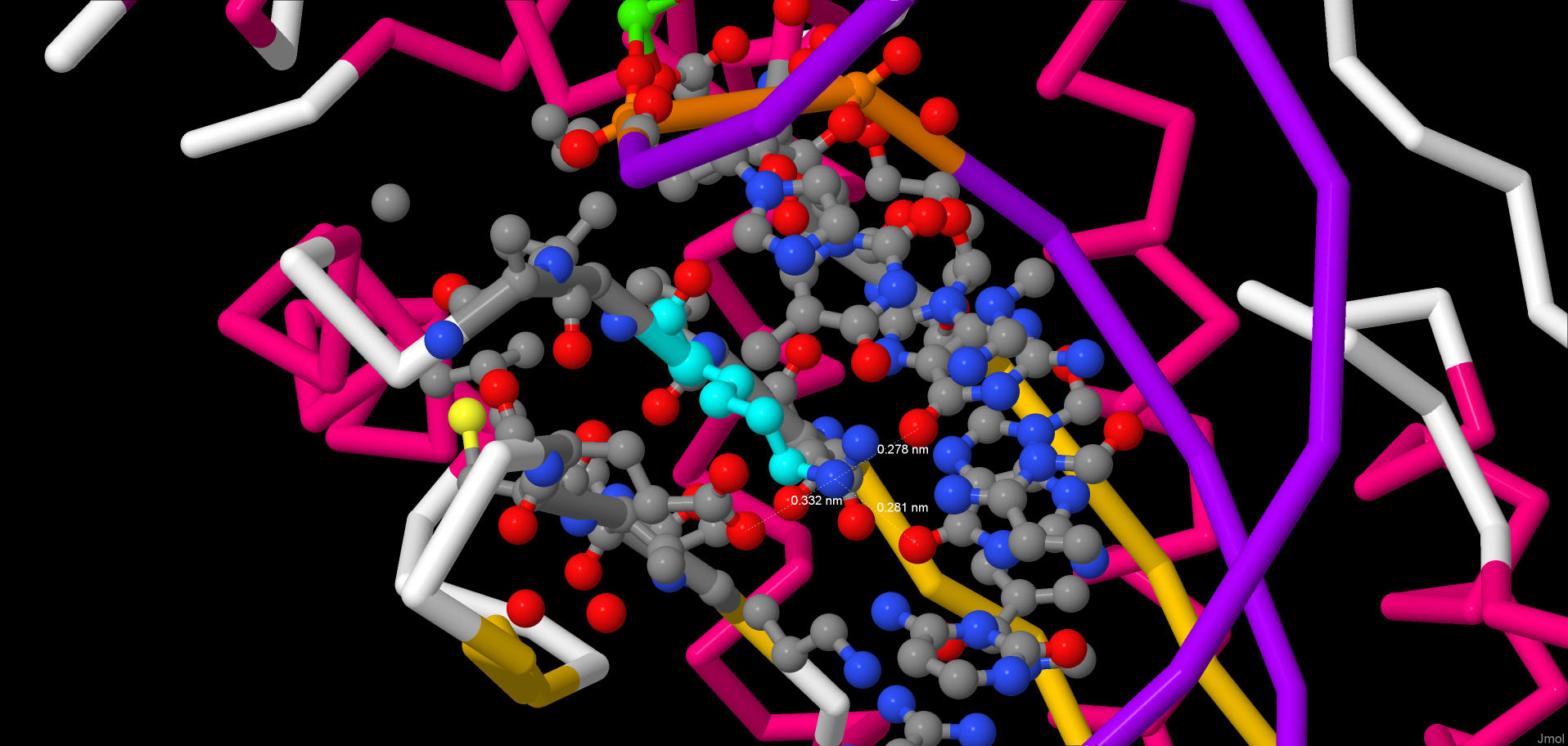 An image of the lysine surrounded by the rest of the structure (the lysine is colored cyan) 2) 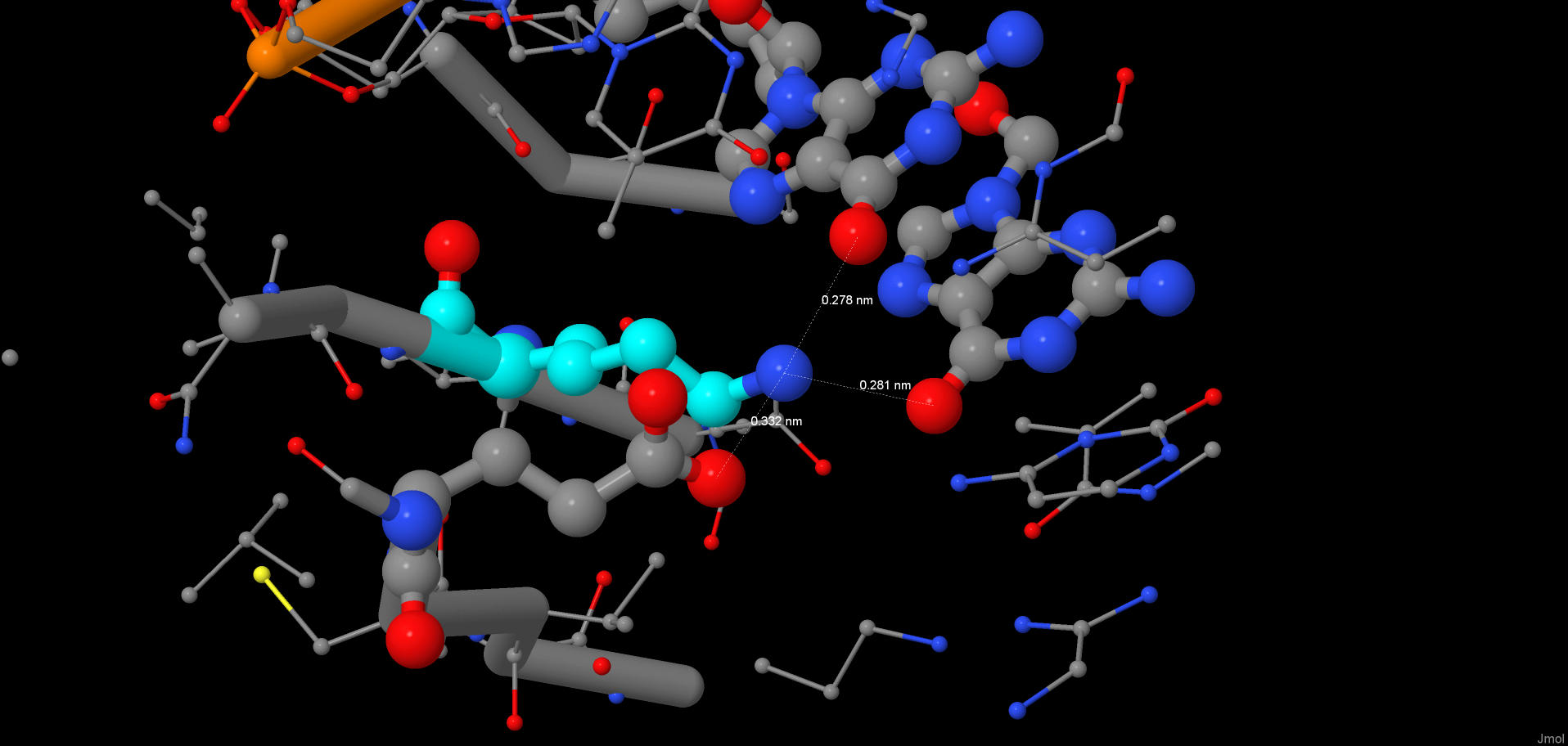 A decluttered version of the previous picture |
© Stanislav Tikhonov, 2018