MALTOPORIN MALTOTRIOSE COMPLEX
Introduction
Maltoporin is a membrane protein, which allows GRAM-negative bacteria, such as Escherichia coli
to transport maltose and maltodextrins across its outer membrane. It is indispensable for the transport of long
oligosaccharides, since they cannot pass through nonspecific porins.[1] Three beta-barrels form the channel of the protein.(Pic. 1)
The sugars are guided into and through the channel construction by hydrophobic interactions with the “greasy slide”-
a hydrophobic path composed of aromatic residues and located at the channel lining.[1,2] The protein is also known as LamB,
as it is misused by a bacterial virus phage Lambda to recognise bacteria.
Uniport ID: P02943
Number of residues: 1263
Chains: A, B ,C (Pic. 1)
Ligands
In our protein magnesium ions bind to the aspartic acid residues (see ligand-protein interactions). Interestingly, in some transporters, such as OmpF channel of E.coli, magnesium binding to the aspartic acid residues increases the affinity of enrofloxacin antibiotic for the channel: the presence of magnisium ion reverses the charge and enrofloxacin may cross the constriction region in its favorable orientation.[4]If the maltotriose concentration is high enough, it is possible for it to pass the membrane not only through LamB, but also through other channels - OmpC and OmpF.[5].
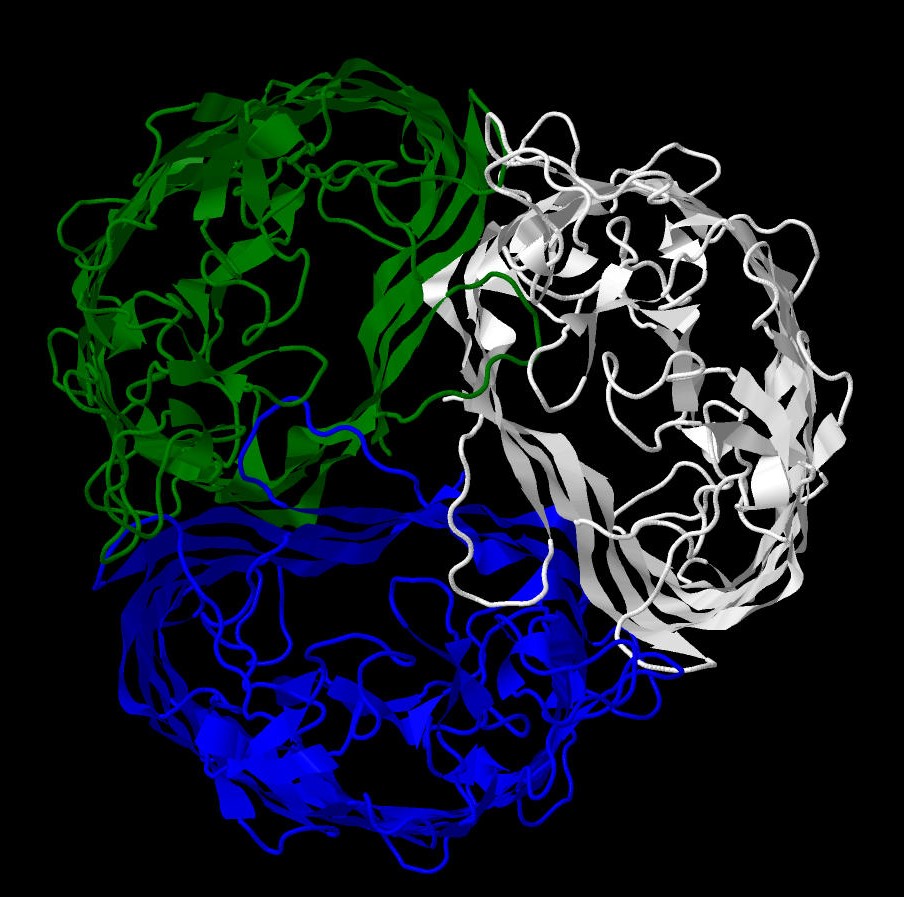
Pic. 1 Protein structure colored by chains

Magnesium ion
Name(IUPAC): Magnesium(2+)
Chemical formula: Mg(2+)
Molar weight: 24.305 g/mol
PubChem ID:888
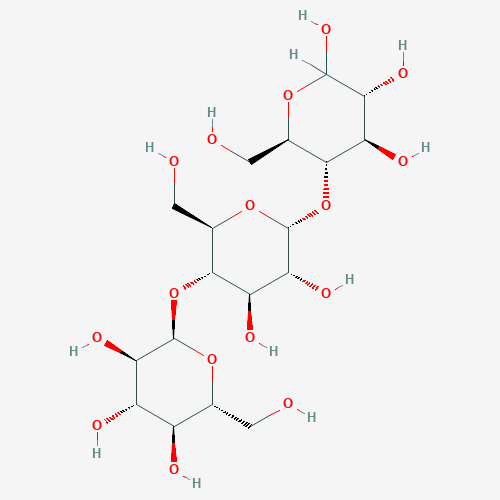
Maltotriose
Name(IUPAC): Alpha-D-gluco-hexopyranosyl-(1->4)-alpha-D-gluco-hexopyranosyl-(1->4)-D-gluco-hexopyranose
Chemical formula: C18H32O16
Molar weight: 504.438 g/mol
PubChem ID:439586
Protein-protein contacts
Hydrophobic interactions play two important roles in a maltoporin molecule: firstly, hydrophobic lining of the channel,
as was previously mentioned, allows oligosaccharides to passively enter the cell; secondly, hydrophobic cores of the
protein form its quaternary structure. It is from this angle that we have considered the hydrophobic core located between
the A and B subunits and formed around a tryptophane residue (TRP101:A). The applet shows the residue and the atoms
surrounding it within the distance of 7 angstrom, but no closer than 1 angstrom. As can be seen from the applet, within
the distance of 3 angstrom there are only atoms, covalently bound to the residue (only 5 atoms), within the distance of 4
angstrom there are 30. From this it can be deduced that the average interatomic distance is about 3.5 angstrom.
Disulphide bridges
Disulfide bridges are the only example of covalent bonds that stabilize the tertiary structure of the protein.
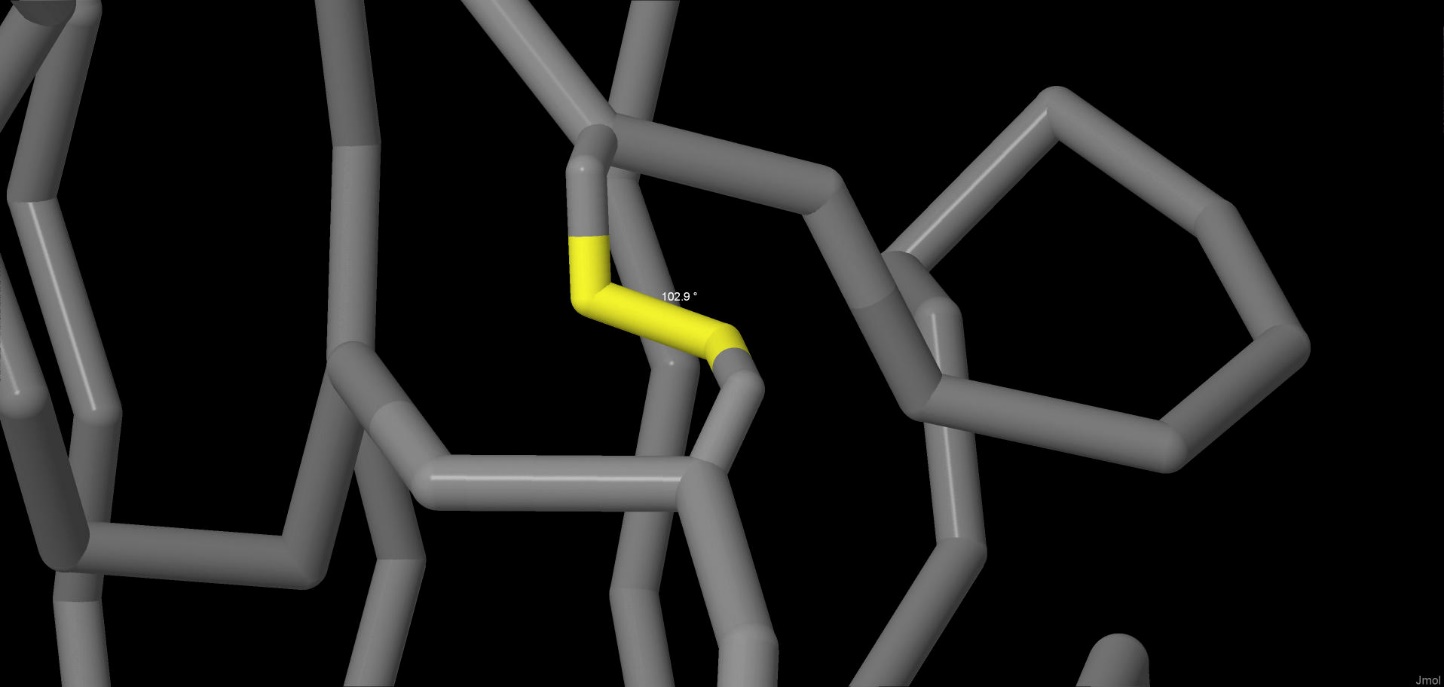
The bond is formed between two cysteine residues. In the molecule there are three disulphide bridges,
one in each subunit. The bridge from the subunit A can be seen in the picture below (CYS022:A– CYS038:A). The measured
torsion angle is 102.9°, whereas the length of the bond is 2.03 angstrom.
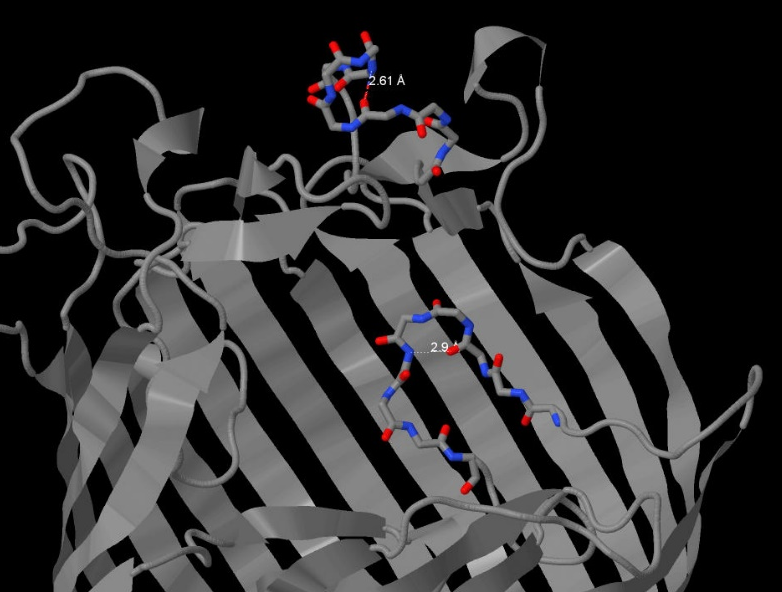
Hydrogen bonds also help the protein form its space structure. The net of hydrogen bonds in maltoporin form a beta-barrel in each subunit, which is very typical for porins. However, we were interested in h-bonds that do not directly take part in forming secondary structure. For example, we have considered 2 hydrogen bonds that form loops in parts of subunit A with primary structure. The length of the bond between the oxygen from carbonyl group in ASN383 and the nitrogen from amine group in ASN387 is 2.61 angstrom, the angle N-O-C is 148.3 angstrom (Pic. 2). The length of a similar bond between the oxygen in THR025 and the nitrogen in ALA028 is 2.9 angstrom, with an angle of 145.4º(Pic. 3). The ideal angle is considered to be 150º, the length of the bond should fall into the range of 2.6-3.3 angstrom.
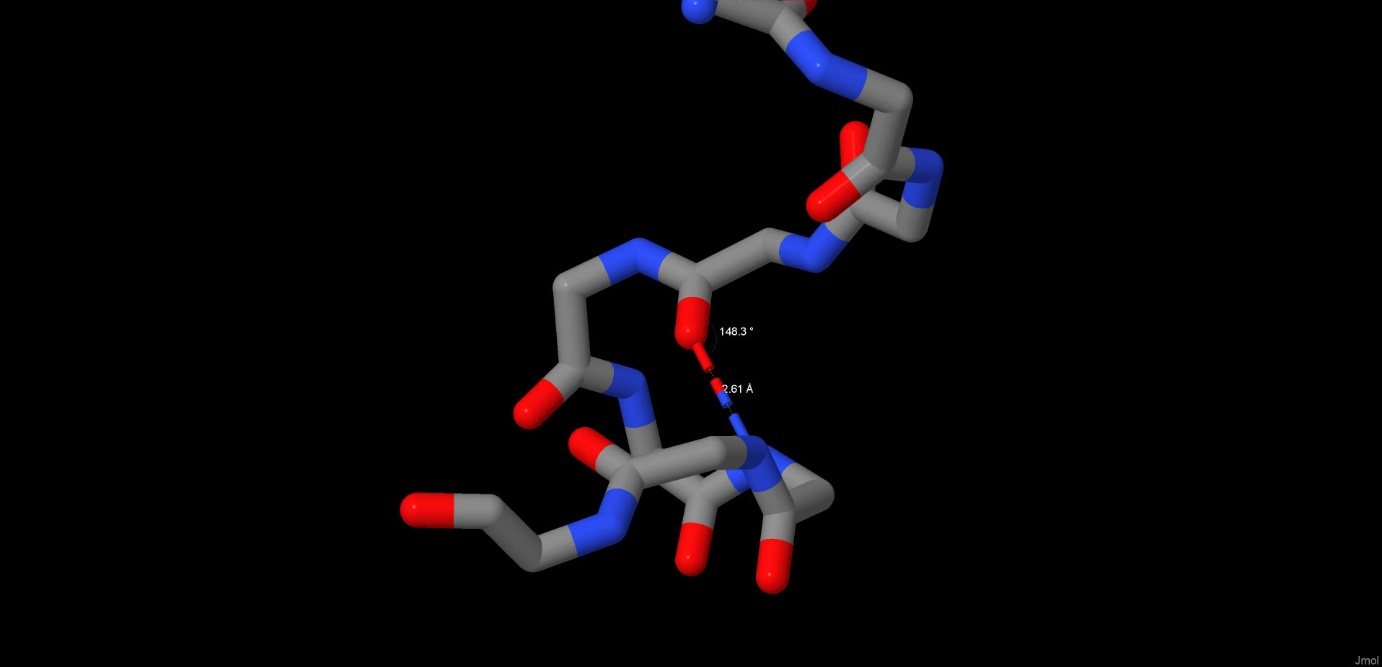
Pic.2 Hydrogen bonds
Beta-barrel
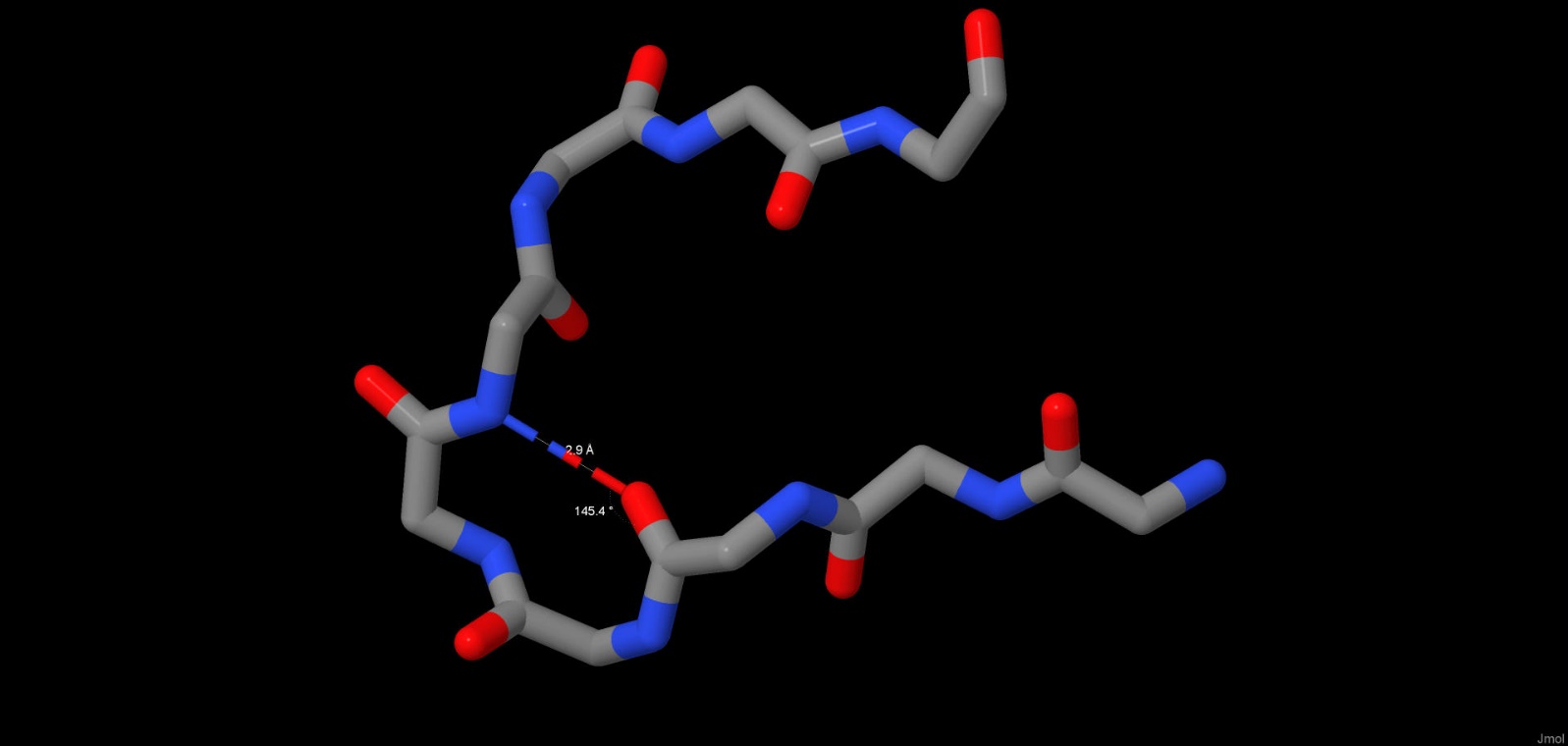
Pic.3 Hydrogen bonds
This type of interactions is electrostatic in its nature. Salt bridges are formed in proteins between the charged parts of a molecule, such as carboxyl group, which is negatively charged at medium and high pH values, and amine group, which is positively charged at the same pH values. Such bonds can be formed with any angles and at a relatively big distance, however they participate in forming the protein structure only if it is under 5 angstrom. The lengths of some of these electrostatic interactions in a malporin molecule can be found in a table below. Salt bridges also help the protein form its tertiary and quaternary structure. The interaction between c and 043GLU:B, 082ARG:B and 073ASP:C can be seen in the picture (Pic. 4).
The salt bridges between the positively and negatively charged residues, whose ionized radicals were orientated in a certain way relative to one another, were found manually. It should also be noted that the distance was measured between the functional groups, whose charges are distributed equally between the atoms, so listing the names of atoms, between which the distance was actually measured, is rather pointless.
| # | A.a.r.(-) | A.a.r.(+) | Bond length, angstrom |
|---|---|---|---|
| 1 | 043GLU | 082ARG | 3.90 |
| 2 | 073ASP:C | 082ARG | 2.70 |
| 3 | 116ASP | 033ARG | 4.02 |
| 4 | 075GLU:C | 106LYS | 3.59 |
| 5 | 039GLU | 039LYS | 5.07 |
| 6 | 061ASP | 045LYS | 2.66 |
| 7 | 188GLU | 110HIS | 3.30 |
| 8 | 218GLU | 110HIS | 3.30 |
| 9 | 002ASP | 004HIS | 4.84 |
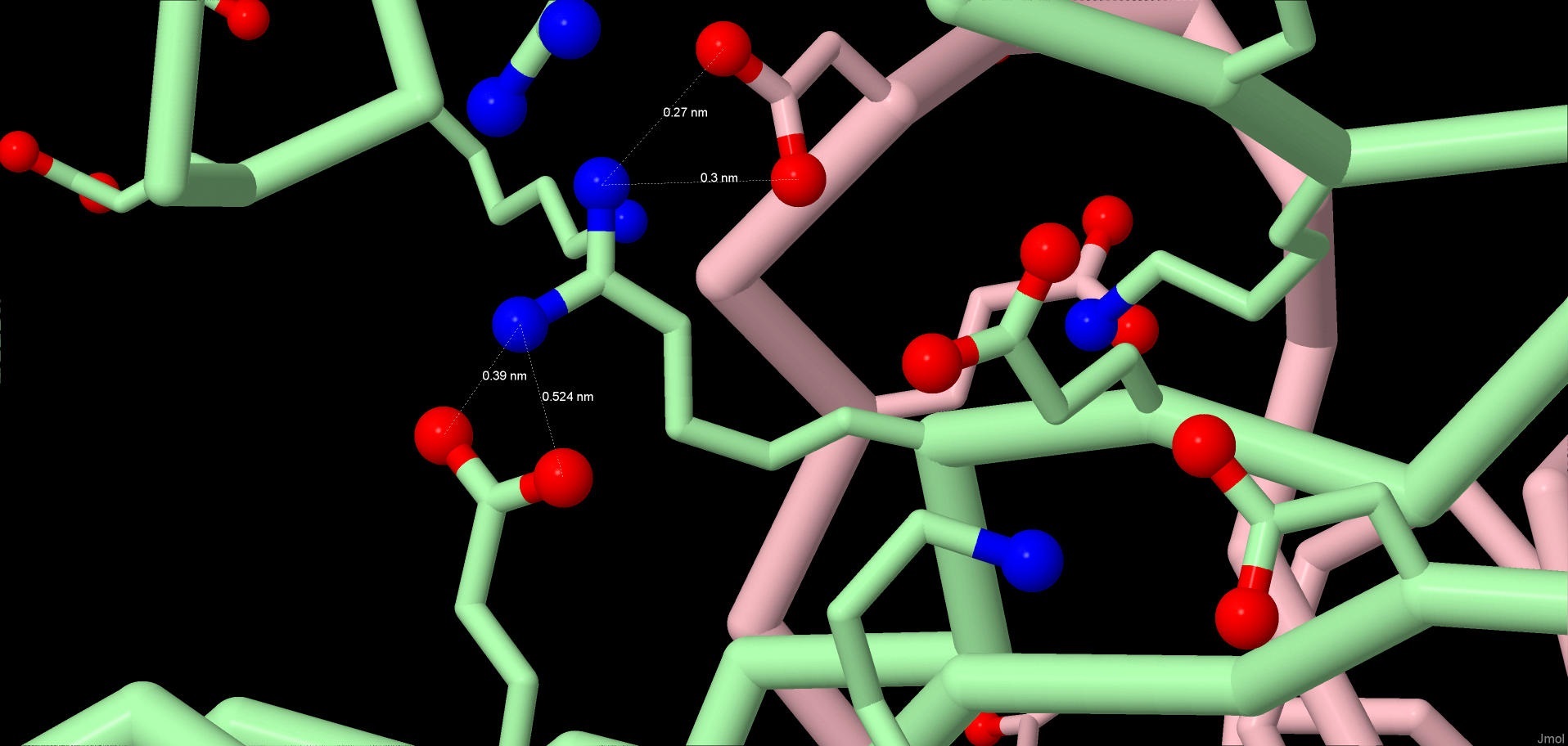
Pic.4 Salt bridges
Ligand-protein interactions
Magnesium ion forms 6 bonds with the protein (for one of the resonance states their median length is 2. 54 angstrom): it interacts with each of the two oxygen atoms of the radicals from the three aspartic acids. (Table 1) Those bonds have already been shown in the protein structure, which was downloaded from PDB. However, we could have predicted their existance without looking at the lines connecting the atoms in the picture. It is well known that the distance between the atoms range between 1 and 4 angstroms [6]. By executing “restrict(4, 422:C)” in Jmol, where 4 is the longest possible distance and 422:C – magnesium ion, we can find all atoms that fall into the 4 angstrom distance of the magnesium atom. What would be left is three aspartic acid residues – positively charged magnisium ion is held in place by the negatively charged carboxyl groups.
| # | Atoms | Bond length, angstrom | |
|---|---|---|---|
| Bonds, formed with [ASP]78:C | |||
| 1 | [MG]422:A - [ASP]78:A.OD1 | 2,52 | |
| 2 | [MG]422:A - [ASP]78:A.OD2 | 3,03 | |
| Bonds, formed with [ASP]78:B | |||
| 4 | [MG]422:A - [ASP]78:B.OD1 | 2,2 | |
| 5 | [MG]422:A - [ASP]78:B.OD2 | 2,56 | |
| Bonds, formed with [ASP]78:C | |||
| 6 | [MG]422:A - [ASP]78:C.OD1 | 2,24 | |
| 7 | [MG]422:A - [ASP]78:C.OD2 | 2,84 | |
| Median | 2,54 | ||
| # | Atoms | Bond length, angstrom | |
|---|---|---|---|
| Bonds formed by [GLC]431:A with the protein atoms | |||
| 1 | [GLC]431:A.O2 [ARG]33:A.NH1 | 2.62 | |
| 2 | [GLC]431:A.O2 [ARG]33:A.NH2 | 3.09 | |
| 3 | [GLC]431:A.O2 [HIS]113:A.ND1 | 3.1 | |
| 4 | [GLC]431:A.O3 [ASP]111:A.OD21 | 3.3 | |
| 5 | [GLC]431:A.O3 [HIS]113:A.ND1 | 2.68 | |
| 6 | [GLC]431:A.O4 [ASP]111:A.OD11 | 3.32 | |
| 7 | [GLC]431:A.O4 [ASP]111:A.OD2 | 3.39 | |
| Bonds formed by [GLC]432:A with the protei atoms | |||
| 8 | [GLC]432:A.O2 [ARG]8:A.NH1 | 3.49 | |
| 9 | [GLC]432:A.O2 [ARG]8:A.NH2 | 3.38 | |
| 10 | [GLC]432:A.O2 [ASP]116:A.OD2 | 2.78 | |
| 11 | [GLC]432:A.O3 [ARG]8:A.NH1 | 3.43 | |
| 12 | [GLC]432:A.O3 [ARG]33:A.NH2 | 2.3 | |
| 13 | [GLC]432:A.O4 [TYR]118:A.OH | 3.49 | |
| 14 | [GLC]432:A.O6 [GLU]43:A.OE1 | 3.08 | |
| 15 | [GLC]432:A.O6 [GLU]43:A.OE2 | 2.52 | |
| 16 | [GLC]432:A.O6 [ARG]109:A.NH2 | 2.93 | |
| Bonds formed by [GLC]433:A with the protein atoms | |||
| 17 | [GLC]433:A.O3 [ASP]116:A.OD2 | 3.13 | |
| 18 | [GLC]433:A.O6 [ARG]109:A.NH2 | 3.16 | |
| Bonds formed between the maltotriose atoms | |||
| 19 | [GLC]431:A.O2 [GLC]432:A.O3 | 2.99 | |
| 20 | [GLC]432:A.O5 [GLC]433:A.O6 | 3.29 | |
| Медиана | 3.115 | ||
Hydrogen bonds between the maltotriose molecules and th protein can be seen not by executing “calculate hbonds”, but on the PDB website, by clicking on «Ligand interaction» in a «Small Molecules» block.
Hydrogen bonds were measured by executing “measure” in Jmol. The median length value is indeed very close to that of the ideal 3.04 angstrem[7]. It should be noted that it would also be possible to find the bonds with a “within” command. For example, if the upper limit of the length of a hydrogen bond were taken as 3.5 angstrem, the result of the “select within(3.5, 431:A)” execution, where 431:A is glucose, would look like this:
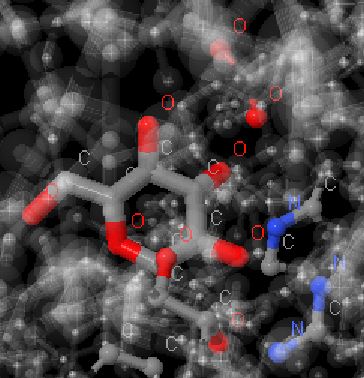
Pic.5. The result of the select within(3.5, 431:A) execution
We can see the same atoms that form hydrogen bonds according to PDB (see applet).
Ptitsina Elena:, scripts and description of the ligand-protein interactions, ligands script
Anna Pago: introduction и ligand descriptions, posting, editing, translation
2. https://www.sigmaaldrich.com/catalog/papers/12003940
3. https://jnanobiotechnology.biomedcentral.com/articles/10.1186/1477-3155-3-3
4. Raj Singh P. et al. Antibiotic permeation across the OmpF channel: modulation of the affinity site in the presence of magnesium //The Journal of Physical Chemistry B. – 2012. – Т. 116. – №. 15. – С. 4433-4438.
5. Лихачева Н.А., Синеокий С.П. Фаговые рецепторы Escherichia coli. Неосновные порины и белки, участвующие в специфическом транспорте как фаговые рецепторы // Молекуляр. генетика, микробиология и вирусология. – 1989. – № 12. – С. 3–12.
6. Ландау Л.Д., Китайгородский А.И. Физика для всех: Молекулы \\6-е изд., стер - Москва: Наука. Главная редакция физико-математической литературы, 1984 – 208 с. [Элекронный ресурс] // Библиотека по физике.
URL: http://physiclib.ru/books/item/f00/s00/z0000016/st008..
7. Стайер Л. Биохимия: Пер. с англ.-М.: Мир, 1984.-Т. 1-232 с., ил.