This protein was obtained from a T. lanuginosa plant by X-ray diffraction.
 Tidestomia lanuginosa
Tidestomia lanuginosa
6HW1-protein belongs to the class of enzymes catalyzing the hydrolysis reactions. So this protein provides splitting of organic compounds with the addition of the water molecule (H + and OH-) elements at the point of rupture.
[2]Here you can see a general view of the reaction catalyzed by hydrolase: A–B + H2O → A–OH + B–H
Lipase is an enzyme that catalyzes the hydrolysis of triglycerides to glycerine and fatty acids. It consists of lipoprotein with inorganic cofactor. Lipases are active surface enzymes. Lipases function on lipid-water section, in such way protein interact with polar and nonpolar molecules.[10] In plants lipases appear in mustard seeds, fruits, tubers, grass rhizome.
.
 |
 |
 |
IUPAC name |
||
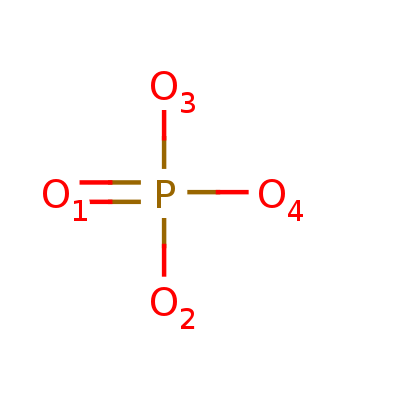
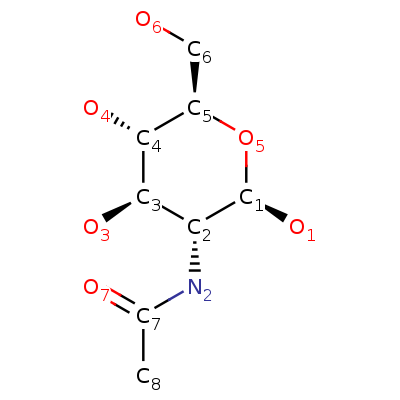
| Magnesium ion | Phosphate ion | β-D-(Acetylamino)-2-deoxy-glucopyranose |
Brutto-formule |
||
| Mg2+ | O4P-3 | C8H15NO6 |
Molar mass |
||
| 24.305 u | 94.97 u | 221.209 u |
PubChem CID |
||
| 888 | 1061 | 24139 |
Ligand PO4 appears only in single protein chain. The rest of ligands dispose symmertrically.
Hydrogen bonds - form of association between an electronegative atom and a hydrogen atom attached to a second, relatively electronegative atom (usually N,O,F).[4] Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). Most H-bonds in proteins appears between N-H и C = O groups in alpha helix and beta sheets. Such bonds we have already represented in Jmol practical work 1, that's why we pay attention on hydrogen bond between [ARG]195:A and [SER]216:A. Distances between nitrogens [ARG]195:A.NE, [ARG]195:A.NH2 and oxygen [SER]216:A.O are common for H-bonds(3.5А).[5] This case shows double hydrogen bonds between aminoacids residues.
The benzene rings of two amino acid residues ([PHE] 7: A and [PHE] 262: A) enter into a T-shaped stacking interaction. The thing is that they are located perpendicular to each other.
More than that, attraction is formed between phenylalanine and tyrosine ([PHE]55:A and [TYR]53:A). This is confirmed by the fact that the aromatic rings are parallel to each other and are at the distance of about 3.5Å, which satisfies the theoretical distance for the formation of pi conjugation.
Ion bridge is one of the interactions, which contributes to the stability to the tertiary structure of protein. It is an electrostatic interaction, arising from the anionic carboxylate ant the cationic ammonium. Asp and Glu are the most common as anion, while His, Arg and Lys are the most common as cation. The distance between the residues participating in the salt bridge is also important and required is less rhan 4 angstrom. To detect ion bridges all pairs of atoms which meet the requirements O-N were highlighted. The green atoms are the atoms of oxygen, the orange atoms are the atoms of nitrogen. Connections and distances are demonstrated in applet.
The contributions of hydrophobic effect[8] to building the tertiary and quaternary structure is great and is about 75 percent. Protein folding is a spontaneous reaction, so it must assume a negative Gibbs free energy value. Water tends to form ordered cages around the non-polar molecule and this leads to a decrease in entropy. With the increase of entropy, Gibbs energy decreases. Minimizing the number of hydrophobic side chains exposed to water is the principal driving force behind the folding process. The multitude of hydrophobic groups interacting within a core of the globular folded protein contributes a significant amount to protein stability after folding.
Hydrophobic cores were found using a service “CluD”[9] to examine the hydrophobic effect. The biggest of them (containing more than 15 atoms) are demonstrated in the applet. Then in the biggest core one of the amino-acid residue(Phe) was chosen and his surroundings was traced. Not a single atom was detected on the distance of one angstrom. Three atoms were the distance of three angstrom, meanwhile on the distance of 4 Å they were found to be 32. Furthermore, when the distance was increase by 1 Å, there appeared to be approximately 30 more atoms in the surroundings.
We can claim that on the distance of 6 Å from the Phe residue there are atoms which completely covet its surface, because with further distancing the core remains as noticeable.
Based on the gathered data we can say that typical distance between neighboring atoms in protein that are not connected covalently comprises around 4-5 Å.
For a water molecule to fit between two neighboring atoms, the average distance between the nuclei of atoms should be more than the sum of their radii and the diameter of water molecule. The smallest radii of all atoms which are included in amino-acids belong to oxygen and nitrogen, that’s why the distance between them should be more than 5.75 Å. But the reasoning cited above show that the average distance approximately equals 4-5 Å, which is considerably less than 5.75 Å. Sp water molecule can not fit between two neighboring atoms.
Sobol Anastasy has written scripts and found information about stacking interactions and hydrogen bonds, designed the html-page and found general information about protein .
Torosyan Tatevik has found the examples of ionic interactions and hydrogen contacts in protein.
Nikolskaya Arina has studied ligands and disulfide bonds; written script that demostartes hydrophobic cores.
Download script: hydrophobic interaction
Download script: ligands
Download script: disulfide bridges
Download script: stacking interaction
Download script: ion bridges
Download script: hydrogen bonds
[6] Spirin А.S. Molecular biology: Ribosome structure and protein biosynthesis, p.256