|
|
|
- Task 1: HMM-profile of the target family
- The motif of the peptidyl-tRNA hydrolase (PTH, PfamAC-PF01195) was chosen, by which was recognised the target family - proteobacteria (phylum proteobacteria)
- During the workshop, an Excel file was created with the "correct" findings of SwissProt, with findings on the HMM profile, a histogram of the weights found and ROC and PRC curves. Alignment used to build an HMM profile. The HMM-profile issued by the hmm2build command
|
|
|
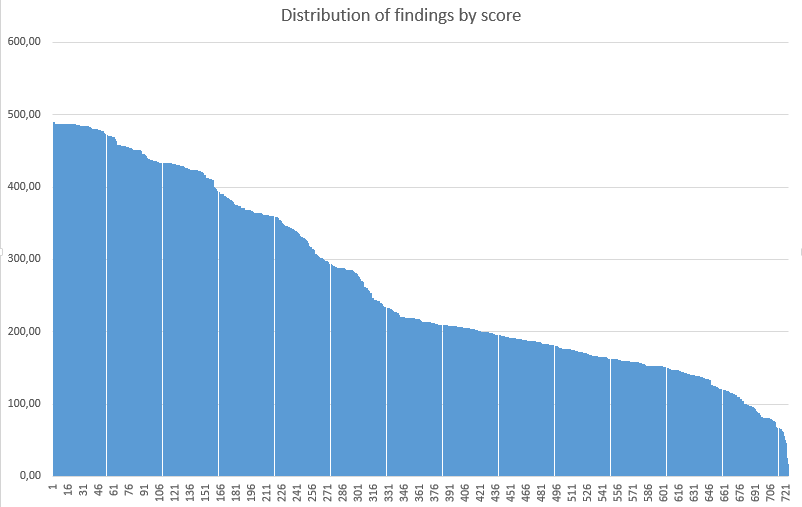
Threshold search:- Histogram of finding's weights
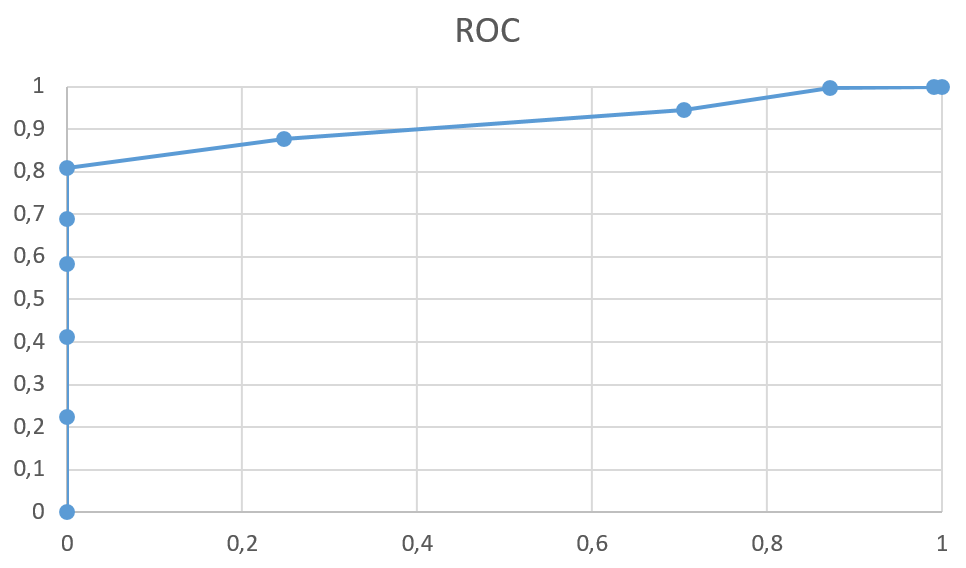
- Roc-curve
- PRC-curve

- From the graphs above, it can be concluded that this method is well suited for finding organisms related to proteobacteria (the ROC curve deviates strongly from the diagonal to the "better" side, like the PRC curve)
The best threshold is 250 or more (score > 250), since the specifity is still 1, and sensitivity is about 81%, there is no way to gain too much, losing less than 20%
- Table with search results at threshold > 250:
|
|
| |
Positive (SwissProt) |
Negative (SwissProt) |
| Positive (predicted) |
314 |
0 |
| Negative (predicted) |
74 |
335 |
| Sensitivity: 0.81 |
Specificity: 1 |
Precision: 1 |
|
|