The overall description of 1XTM structure
- PDB identification code: 1XTM
- Molecule: hypothetical superoxide dismutase-like protein yojM
- There are 2 chains (A,B) corresponding different protein forms (Y88H and P104H) in the PDB-file
- The PDB-file also contains the coordinates of the following ligands:
- In the A-chain there is 1 copper-ion Cu2+ (503) and 1 zinc-ion Zn2+ (504)
- Inthe B-chain there is 1 copper-ion Cu2+ (500) and 4 zinc-ion Zn2+ (501,502,505,506)
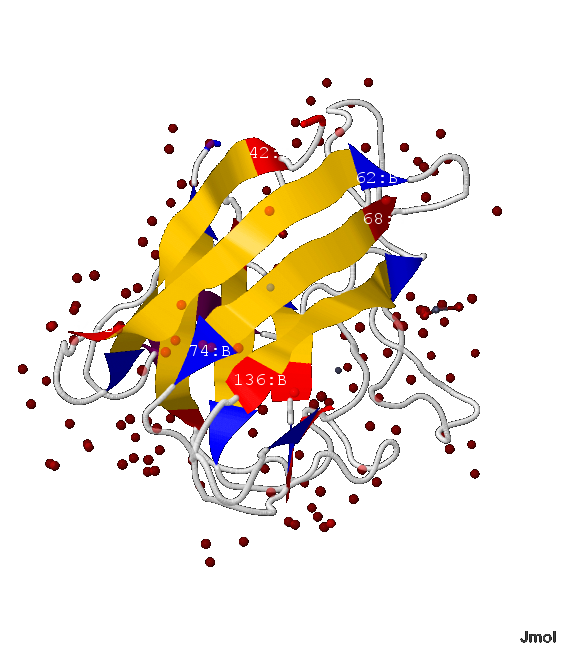
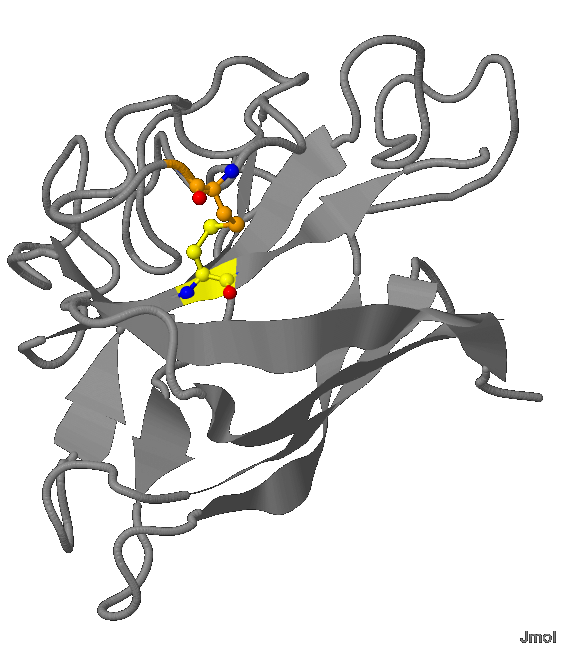
Secondary structure of the protein is shown in fig. 1
.png) Fig.1 The protein structure. The chain movement.
Fig.1 The protein structure. The chain movement. Common: copper atoms are bronze (color), zinc atoms - grey; water molecule - red; β-sheets are presented in the form of ribbons; white threads - the rest of the protein
The movement of the protein chain is shown by the sequence of colors:
Chain А: maroon N-end » β-strands: red » scarlet » pumpkin » orange » amber » yellow » bright green » green » Cal Poly green » fern green C-end
Chain B: aquamarine N-end » β-strands: cyan » azure » cobalt blue » blue » ultramarine » indigo helix » β-strands: violet » magenta » coral pink » orchid » wisteria C-end
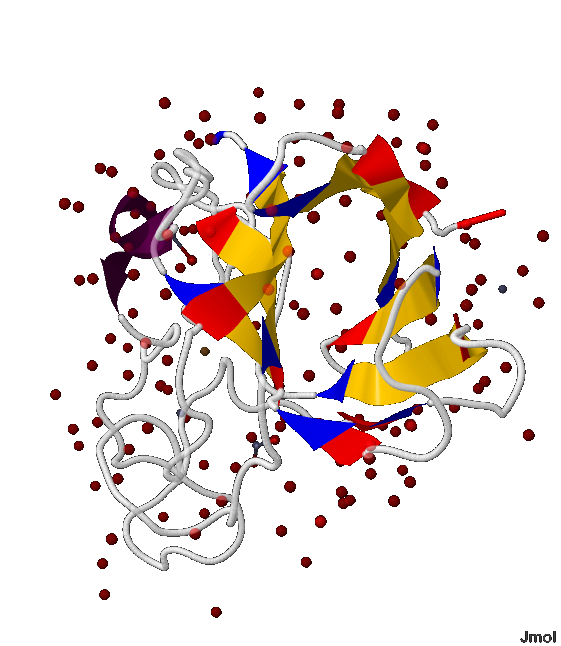
Structure analysis of helices and beta-sheets in the chain B of the protein with 1XTM PDB ID
In 1XTM protein the B-chain contains both the helix and β-sheet that is why I have chosen it for the secondary protein structure analysis.
In picture 2 the location of the helix and β-sheet in the protein can be clearly seen (the helix is colored in violet and β-sheet is colored in yellow)
The helix analysis
In 1XTM protein the helix starts with the 98th chain-B amino acid residue and finishes at the 102nd.
Constitutive properties:
- The helix consists of 5 aminoacid remainders (1,67 of the spire);
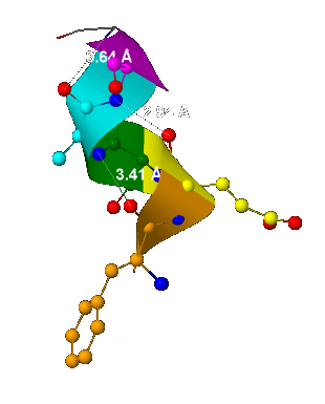
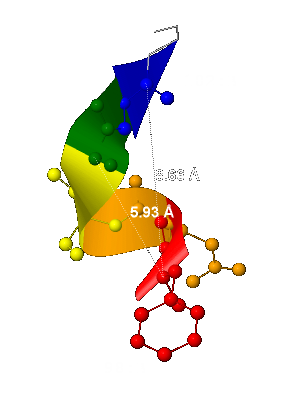
- The helix pitch - 5,93 Å
(it was computed as the distance between 2 the nearest and located one above the other carbon α-atoms – fig.3); - The number of the helix residue for 1 turn is 3.
(It can be seen in fig.3: 3 aminoacid residue – red, orange and yellow – form 1 spire); - The number n residue forms hydrogen bond with n+3 residue
(some of the hydrogen bonds are shown as dotted line in fig.4)
| Geometry attribute | α-helix | 310-helix | π-helix | 1XTM-helix |
| Residues per turn | 3,6 | 3,0 | 4,4 | 3 |
| Pitch | 5,4 Å | 6,0 Å | 4,8 Å | 5,93 Å |
Constitutive properties of 3 main protein helices forms and 1XTM protein helix data are tabulated in table1. While comparing them one can conclude the 1XTM helix is 310-helix.
β-sheet analysis
In 1XTM protein there is β-sheet consisting of 9 β-strands: 42-49:B, 54-62:B, 68-74:B, 82-83:B, 85-88:B, 128-129:B, 136-142:B, 161-166:B, 183-189:B amino acid residue.
- In this β-sheet there are 2 parallel β-strands - 82-83:B and the subsequent 85-88:B; and all the rest are antiparallel , т.е. напротив N-начала каждого β-тяжа расположен C-конец последующего и предыдущего (показано на рис.5-7)
- Данный β-лист образует почти замнкнутую объёмную фигуру, напоминающую бочонок (показано на рис.5-8)
Внутримолекулярные взаимодействия боковых групп белка в цепи A структуры 1XTM
Так как в предыдущем разделе рассматривалась цепь B белка 1XTM, то при анализе взаимодейсвия боковых групп белка я решил изучить цепь A.
- В белке 1XTM содержится 2 цистеиновых остатка (93 и 186), связанных дисульфидным мостиком (показано на рис.9)
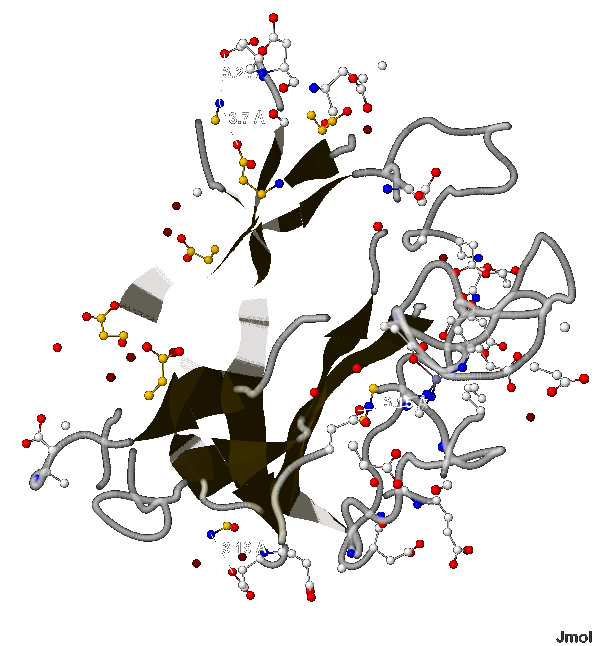
- Согласно [2] длина соляного мостика не может быть больше 4 Å. В А-цепи белка 1XTM мною найдено 4 таких связи (изображены на рис.10) длиной:
- 3.19 Å (между 49-ым и 51-ым аминокислотными остатками аспарагина и глутамата соответственно),
- 3.15 Å (между 112-ым и 170-ым - гистидина и аспартата),
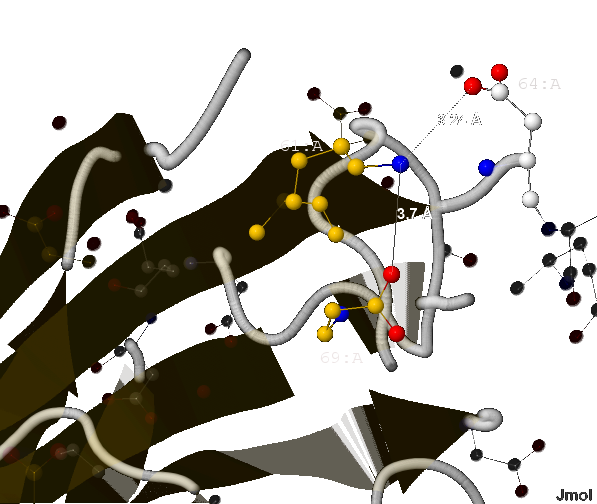
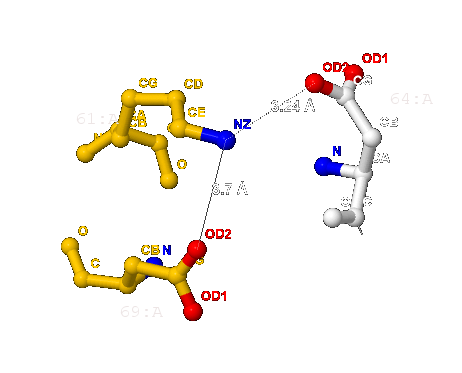
- 3.7 Å (между 61-ым и 69-ым - лизина и аспартата) и
- 3.24 Å (между 61-ым и 64-ым - лизина и тоже аспартата);