| FBB MSU site | Main page | About me | Terms |
Domains. Pfam.
Task 1
Domain choise
In database Pfam I chose a family of domains: Class II histocompatibility antigen, beta domain (MHC_II_beta).
AC: PF00969
ID: MHC_II_beta
10 architectures
74 species
600 sequences
228 structures
Below you can see 3D structure of human MHC class II molecule (beta chain) in complex with staphylococcal enterotoxin I (SEI) (fig. 1).
MHC class II is an antigen-presenting molecule involved in adaptive immune response generation. It is recognized by C4 or C8 T-cell receptors. MHC class II molecule is a heterodimer, it include alpha and beta domains. These domains form an intramembrane compartment and antigen-presenting groove on the outside of the membrane.
PDB ID: 2G9H
Classification: IMMUNE SYSTEM
Organisms: Homo sapiens, Influenza A virus (strain A/England/878/1969 H3N2), Staphylococcus aureus
Expression System: Escherichia coli
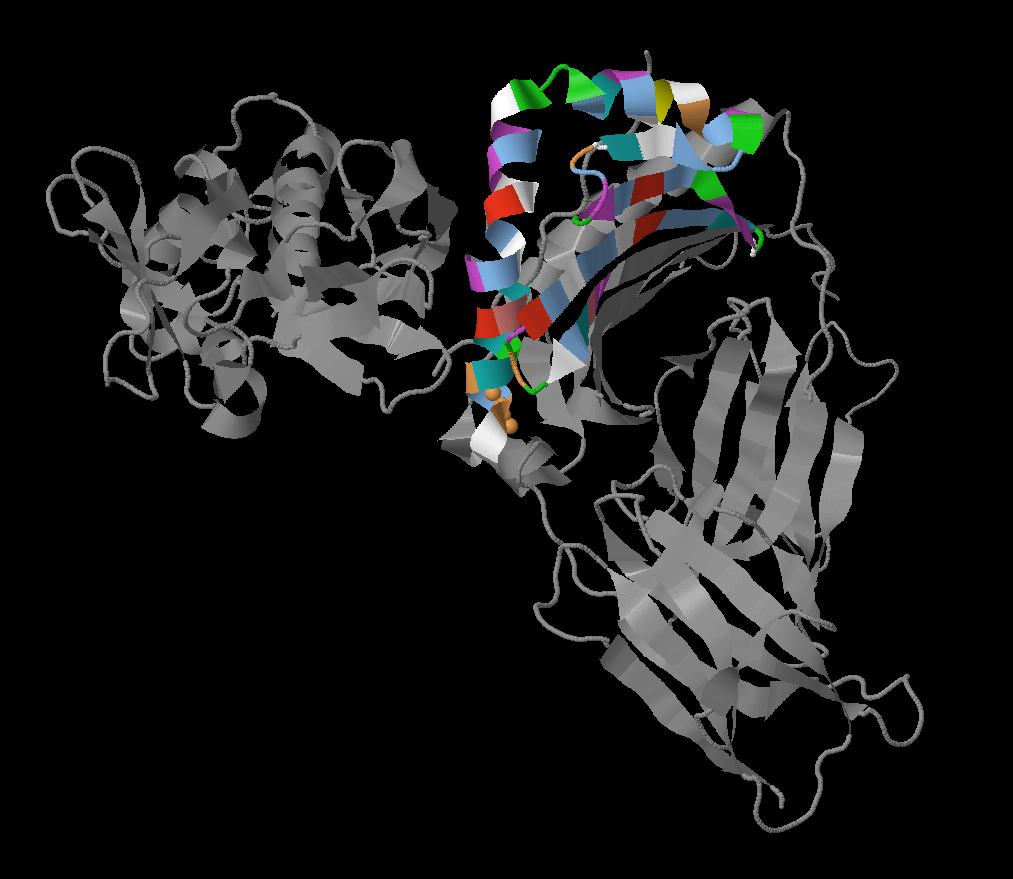
Figure 1. Structure of Staphylococcal Enterotoxin I (SEI) in Complex with a Human MHC class II Molecule.
Jalview project of all sequences aligned: MHC_II_beta_align_all.jar.
Architectures choise
There are 492 sequences with the following architecture: MHC_II_beta, C1-set
Further this architecture will be called "due".

Figure 2. MHC_II_beta, C1-set.
There are 87 sequences with the following architecture: MHC_II_beta
Further this architecture will be called "uno".

Figure 3. MHC_II_beta.
Using script I receive information about all sequences belonging to Pfam family.
python swisspfam-to-xls.py -i /srv/databases/pfam/swisspfam.gz -z -p PF00969 -o PF00969.xls
Summary table
Using ВПР (LibreOffice) I formed a summary table where rows are AC-s and columns are Pfam domains. You can see it here: PF00969+taxonomy.ods.
Taxons choise
Firstly I saved a list of AC-s from PF00969.xls to ac.txt. Then I get the whole sequences of these proteins by Uniprot-Retrieve searching and then using script I receive information about taxonomy of saved MHCIIb sequences. Then data was added to the previous table.
python uniprot_to_taxonomy.py -i uniprot-yourlist.txt taxonomy_only.xls
Analyzed data I found out that all sequences belong to taxon Euteleostomi (Eukaryota, Metazoa, Chordata, Craniata, Vertebrata). This taxon include two enough numerous subtaxons: Archelosauria (56) and Mammalia (387).
Taxon - Euteleostomi
Subtaxons - Mammalia (M), Archelosauria (A)
Architectures - uno, due
Representatives choise
I chose 17 represetatives of due in each subtoxons and 14 represetatives of uno per subtaxon. Their AC-s were saved as ac_chosen.txt.
Final table
The resulted table is here: PF00969+taxonomy.ods.
Reseived files were converten into spreadsheet. File PF00969+taxonomy.ods contain sheets: PF00969 (received by script), summary_ table (rows - AC, columns - Pfam domains), taxonomy (received by script), chosen sequences (of subtaxons) (include one with known 3D structure), final table (all useful information about sequences). All that applies to uno are colored light pink and to due are colored light green.
Only the selected sequences were left in the basic alignment (MHC_II_beta_align_all.jar) by script (you can see command below).
python filter-alignment.py -i MHC_II_beta_align_all.fasta -m ac_chosen.txt -o chosen_align.fasta -a "_"
Sequences in reseived 'filtered' alignment were renamed by principle: architecture_subtaxon_AC.
Reference designations:
- uno - MHC_II_beta
- due - MHC_II_beta, C1-set
- M - Mammalia
- A - Archelosauria
Then they were sorted by ID. Empty columns, N-end and C-end sections were deleted. Bad aligned sequences were deleted too. Different architectures were separately grouped and colored by ClustalX by conservation (50). Final project you can see follow this link - chosen_align.jar and in figure 4.
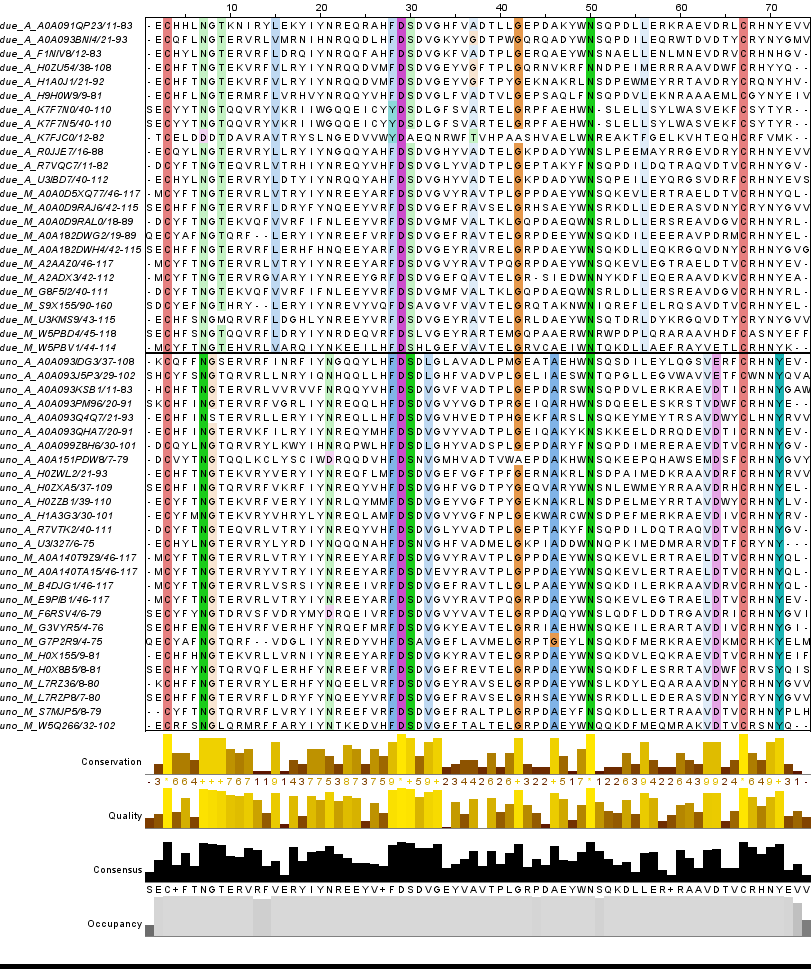
Figure 4. Final alignment of chosen representatives.
Task 2
Phylogenetic tree of the domain
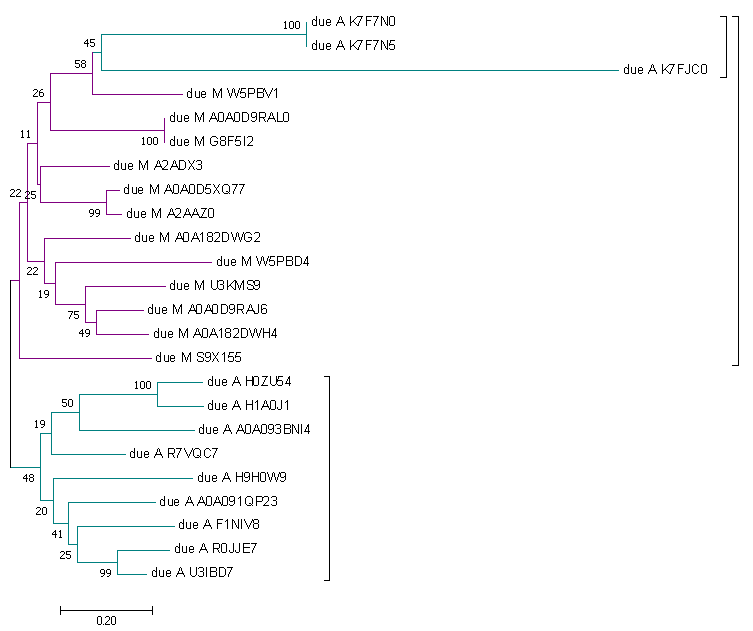
To bild a tree I used Neighbor-Joining, Bootstrap 100 (MEGA). The phylogenetic tree of MHC_II_beta domain you can see below. Trees for 'due' (fig. 5) and 'uno' (fig. 6) were builded separately.
Bracket tree formulae:
 |
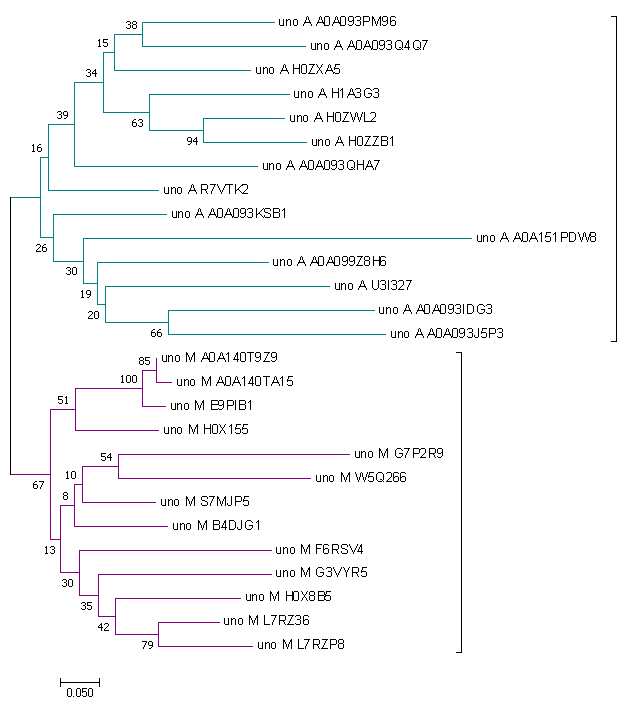 |
| Figure 5. Tree of representatives of architecture due. | Figure 6. Tree of representatives of architecture uno. |
Conclusion:
Analizing the uno phylogenetic tree we can clearly see divergence of Mammalia and Archelosauria subtrees. So I guess that last common ancestor of these groups had this architecture before their divergence.
As for the due phylogenetic tree we notice clearly divergence too but there are a subtree of Archelosauria inside the global Mammalia subtree. This situation most likely is the result of an unsuccessful choice of representatives or a program error. Nevertheless I can assume some possible reasons for such an evolutionary event. For example, this domain architecture could had appear in these taxons independently. Or it might be a result of a later separation of this small group of Archelosauria from the general tree. But this version is very doubtful because it isn't verified by uno tree.
| Term 4 |
| ← Pr 6 | → Pr 8 |
© Darya Potanina, 2017