| FBB MSU site | Main page | About me | Terms |
Genomic environment
Database GO
COG for FBPase
My protein: fructose 1,6-bisphosphatase - FBPase - ALX49872.1 (descroption; fasta).
COG (Clusters of Ortologous Groups of proteins) search:
Program: CDD (Conserved Domain Database)
Result:
- COG ID: COG1494
- E-value: 0e+00
- Interval: 1-320 (protein lenght 322)
- COG name: Fructose-1,6-bisphosphatase/sedoheptulose 1,7-bisphosphatase or related protein
- Function group: Carbohydrate transport and metabolism
Visualisation of genomic environment of FBPase
Gene Neighborhood Analysis:
Program: COGNAT (COmparative Gene Neighborhood Analysis Tool)
- Search for COG: COG1494
- Neighborhood Size: 9
- Occurrence Threshold: 10% (OT > 15% didn't find any neighboring genes)
- Taxonomy: No
Result you can see below.

Figure 1. The first group of neighboring genes.
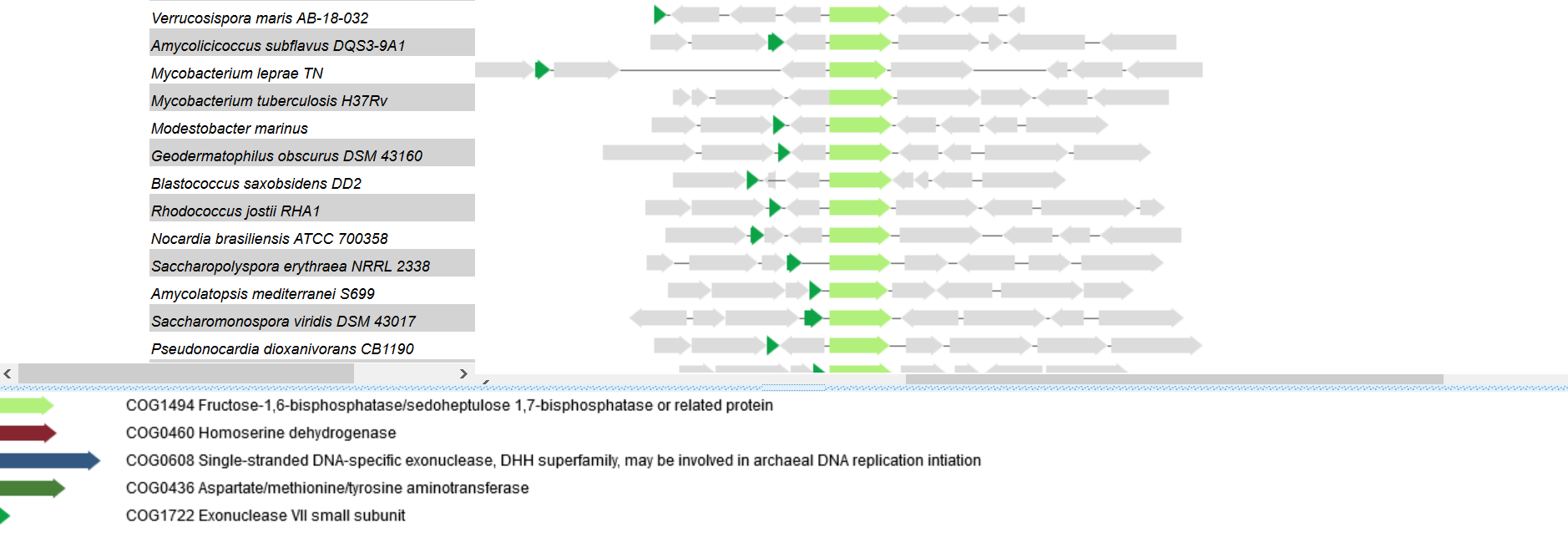
Figure 2. The second group of neighboring genes.
Neighboring genes don't relate to fructose-1,6-bisphosphate metabolism, so I think there isn't static genomic environment. Bacteria have similar genomic environment of Fructose-1,6-bisphosphatase gene belong to Alphaproteobacteria (fig 1) or Actinobacteria (fig 2). So this architecture can be called as "linked genes", but it isn't a group of genes coding macromolecular complex.
FBPase in GO terms
The FBPase protein was quieried with AmiGO BLAST against its database to find the most similar available protein. The best finding has p-value of 1.9e-124; it is a fructose-1,6-bisphosphatase, class II (BA_5576; Uniprot: Q81JW7; Pfam: PF03320) from Bacillus anthracis str. Ames. I think GO terms can be associated with FBPase protein (see table 1).
Table 1. Go terms related to Q81JW7.
| Point | GO ID | Term | Evidence |
| Biological process | GO:0030388 | Fructose 1,6-bisphosphate metabolic process | ISS |
| Biological process | GO:0006094 | Gluconeogenesis | ISS |
| Molecular function | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity | ISS |
ISS means, that Inferred from Sequence or structural Similarity i.e. from manual analysis of genome annotation or from structural similarity with experimentally characterized gene products.
| Term 4 |
| ← Pr10 | Pr 12 → |
© Darya Potanina, 2018