Atlas of contacts PKG I's Carboyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP
Introduction
|
PKG (cGMP-dependent protein kinase or Protein Kinase G) is a serine/threonine-specific protein kinase that is activated
by cGMP [1] ((Cyclic guanosine monophosphate) is a cyclic nucleotide derived from guanosine triphosphate (GTP) [2]).
Serine/threonine protein kinase that acts as key mediator of the nitric oxide (NO)/cGMP signaling pathway.
GMP binding activates PRKG1, which phosphorylates serines and threonines on many cellular proteins. Numerous protein
targets for PRKG1 phosphorylation are implicated in modulating cellular calcium, but the contribution of each of these
targets may vary substantially among cell types.
PKG phosphorylates a number of biologically important targets and is implicated in the regulation of smooth muscle
relaxation, platelet function, sperm metabolism, cell division, and nucleic acid synthesis. [3]
|
General information:
•PDB ID: 5J48
•Uniprot ID: Q13976
•Chains: A, B (Fig1)
•Sequence Length: 135
•Organism: Homo sapiens
•Gene Names: PRKG1 (PRKG1B, PRKGR1A, PRKGR1B) EC: 2.7.11.12
|
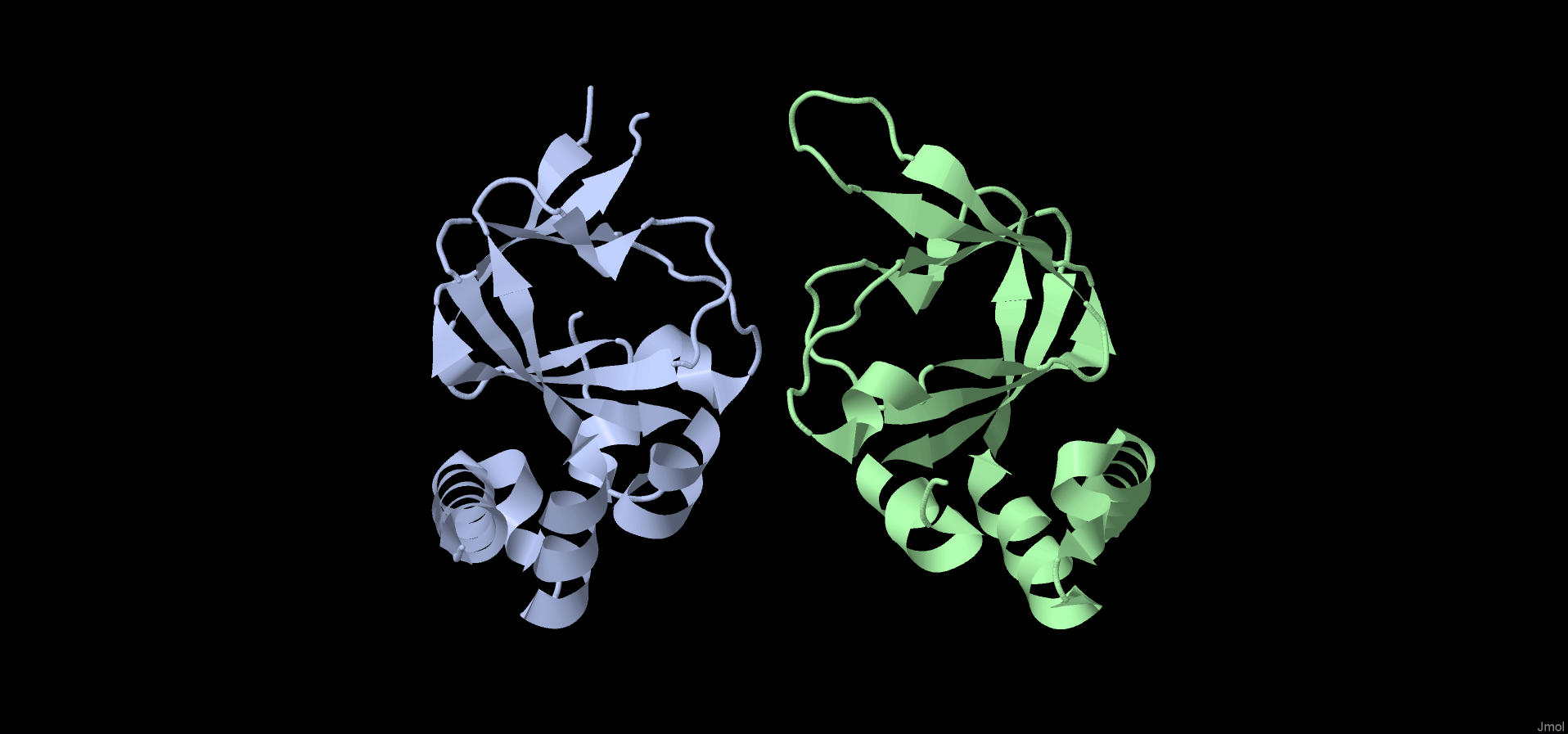 Fig.1. Protein structure colored by chains. Chain A is blue. Chain B is green.
Fig.1. Protein structure colored by chains. Chain A is blue. Chain B is green.
|
There are 4 ligands in the biomolecule: |
| 6FW | NA (SODIUM ION) |
|---|
IUPAC Name: 9-[(4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxo-4a,6,7,7a-tetrahydro-4H-furo[3,2-d]
[1,3,2]dioxaphosphinin-6-yl]-2-amino-8-(4-chlorophenyl)sulfanyl-3H-purin-6-one
Structure: Fig.2
Molecular Formula: C16H15ClN5O7PS
Molecular Weight: 487.808 g/mol
PubChem ID:123969
|
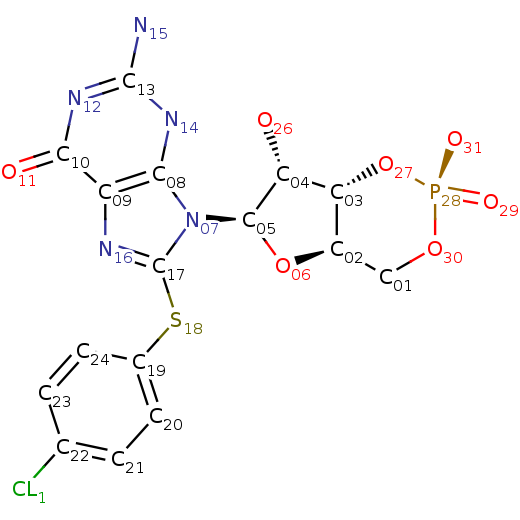 Fig.2. Structure of 8-Pcpt-cgmp.
Fig.2. Structure of 8-Pcpt-cgmp. |
IUPAC Name: sodium(1+)
Structure: Fig.3
Molecular Formula: Na+
Molecular Weight: 22.99 g/mol
PubChem ID:923
|
 Fig.3. Sodium ion.
Fig.3. Sodium ion. |
| Ca (CALCIUM ION) | 1,2-ETHANEDIOL (ETHYLENE GLYCOL) |
|---|
IUPAC Name: calcium(2+)
Structure: Fig.4
Molecular Formula: Ca+2
Molecular Weight: 40.078 g/mol
PubChem ID:271
|
 Fig.4. Calcium ion.
Fig.4. Calcium ion. |
IUPAC Name: ethane-1,2-diol
Structure: Fig.5
Molecular Formula: C2H6O2
Molecular Weight: 62.068 g/mol
PubChem ID:174
|
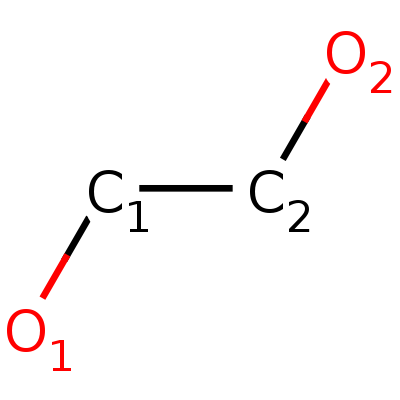 Fig.5. Structure of ethylene glycol.
Fig.5. Structure of ethylene glycol. |
|
Protein-protein contacts
Salt bridge (or ion bridge) is a combination of two noncovalent interactions: hydrogen bonding and
ionic bonding. The salt bridge most often arises from the anionic carboxylate (RCOO-) of either
aspartic acid or glutamic acid and the cationic ammonium (RNH3+) from lysine or the guanidinium (RNHC(NH2)2+) of arginine. [4]
Command “Restrict positive or negative” was used for searching salt bridges. Then corresponding contacts were searched manually.
After that bond distance were measured. If bond length was less then 4.0 A, bond was considered salt bridge.
Two chains of protein are connected to each other by ion bridge (Fig.6). May be, this type of interaction is pretty strong.
Another salt bridges are represented in the applet. They are inside the same chain.
|
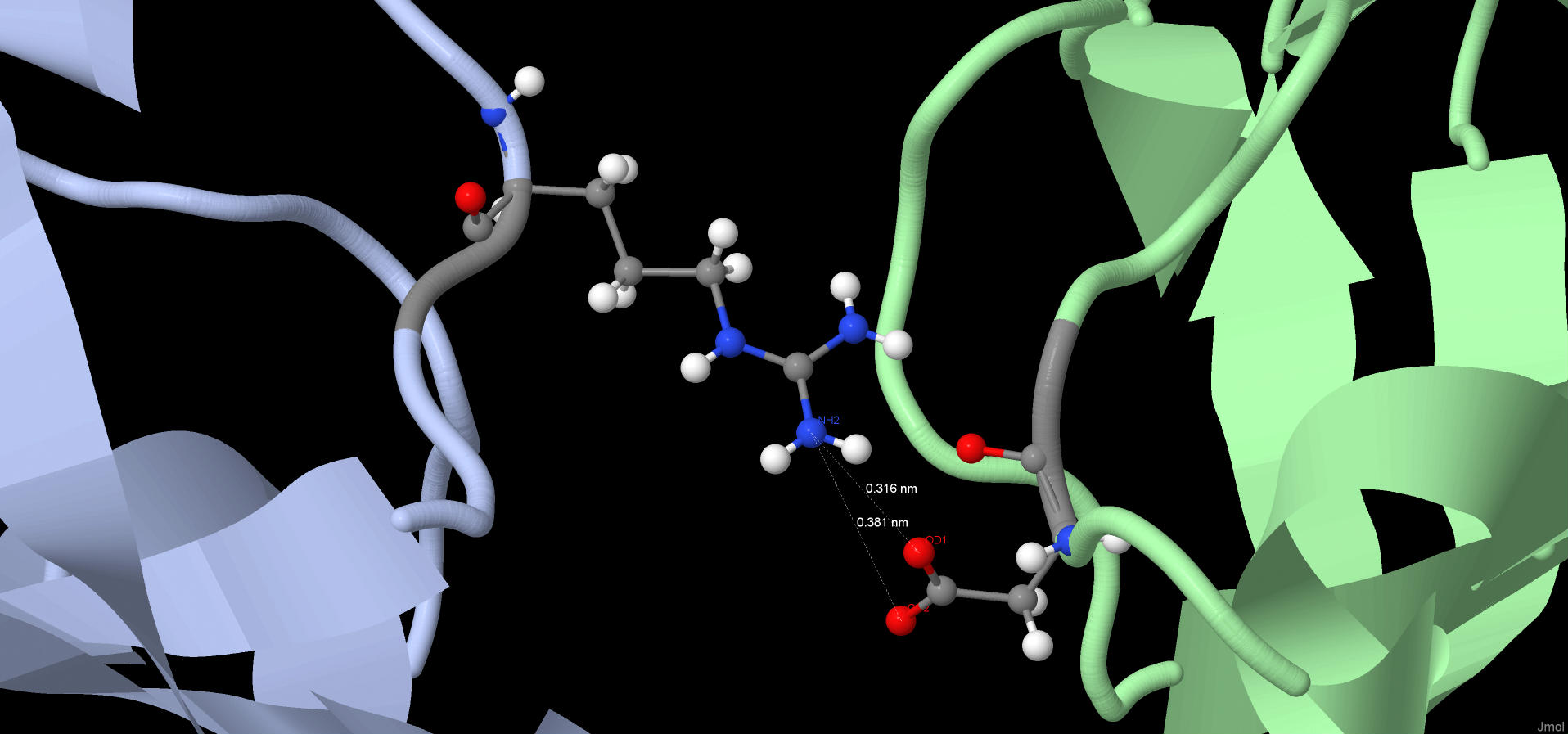 Fig.6. Salt bridge connected different chains of protein.
Chain A is blue. Chain B is green.
Red balls are oxygen. Blue balls are nitrogen.
Fig.6. Salt bridge connected different chains of protein.
Chain A is blue. Chain B is green.
Red balls are oxygen. Blue balls are nitrogen.
|
|
According to the IUPAC definition, “The hydrogen bond is an attractive interaction
between a hydrogen atom from a molecule or a molecular fragment X–H in which X is more electronegative than H,
and an atom or a group of atoms in the same or a different molecule, in which there is evidence of bond formation” [5].
We discussed hydrogen bonds at the seminar on bioinformatics. And we decided that distance of hydrogen bond
is from 2 to 3.5 A and the angle should be as close as possible to 180 °.
Command "hbonds calculate" was used for the searching of hydrogen bonds.
A total of 466 hydrogen bonds were found in the biomolecule, of which 303 are hydrogen bonds within the protein
(with a length of the protein of 135 amino acid residues). We assume that such a large number of bonds
(2.2 times greater than the amino acid residues) is necessary to stabilize the quaternary structure of the protein.
It also turned out that Jmol, considered salt bridges hydrogen bond, which is partly true, but not quite so,
it is necessary to take into account the contribution of ionic interaction to the formation of salt bridges.
When we used command "hbonds calculate", ion bridges have been defined as hydrogen bonds. And this means that the number
of hydrogen bonds in the biomolecule calculated by the program is less than it actually is.
The applet represents hydrogen bonds between the beta-sheets and within single alpha-helix.
|
|
|
Covalent bonds |
Covalent bonds are represented by peptide bonds. A coordination bond between sodium and two amino acid residues,
glutamine and lysine, is also found, which is presented in the applet. In this case, metal ions interact with the protein. |
Packing density of hydrophobic core |
|
The hydrophobic core is a region of space with a high density of side chains of hydrophobic amino acids.
Hydrophobic interactions have a key role in folding the protein globule. Hydrophobic interactions are entropic in nature,
since the formation of contacts between water and nonpolar molecules is thermodynamically unprofitable. [6]
The search for a hydrophobic core to investigate the density of its packaging was carried out manually.
First, large hydrophobic amino acids (in this case phenylalanine) were isolated. Next, the biomolecule was examined and the
hydrophobic core was determined. The hydrophobic core represented in the applet is located approximately in the center of the B chain.
And it appears that it forms the quaternary structure of the protein.
The minimum distance from the residue on which the atoms are located that completely cover the surface of the hydrophobic
core is approximately 5-6 A. For atoms that are not covalently bound, a distance of 4.2-5 A is characteristic. Two atoms were randomly
selected. It was visually close to each other. Further, several tens of distances were measured manually. After that, the average range was taken.
Between neighboring atoms can not fit another, because The minimum necessary distance for a molecule of water,
which is essentially one patient oxygen, is 2.8 A. And the distances between neighboring atoms are considered from the center,
without taking into account the radius.
|
|
|
|
Salt bridges |
| One of the ligands is bound to the protein by three salt bridges. All of them are presented in the applet.
Perhaps this is due to the fact that this ligand performs an important function. |
Hydrogen bonds |
|
29 hydrogen bonds were found between the protein and ligands. The "hbonds calculate" command was used for calculating
their number, before that protein and ligands were isolated and water, which is not a ligand, is formed,
but forms hydrogen bonds. The applet represents the hydrogen bond between glutamine in the protein and ethylene glycol,
which is a ligand. This ligand is bound to the protein by only one hydrogen bond, it is possible that it does
not play an important role, although the opposite is not excluded. This is just a guess.
|
|
|
|
|