Contact atlas
Hydrogen bonds
Hydrogen bonds retain secondary structure of protein and they can help forming 310-helix, where i-aminoacid connects with i+3 aminoacid in contrast to alpha helix, where I connects with i+4. The 310-helix is usually 2 turns long and it is coloured in magenta when color structure Jmol-command is used.
Length of hydrogen bonds in such a structure: [SER]209:F-[GLY]206:F 4.27Å, [GLU]101:C-[ILE]104:C 3.22Å, [HIS]415:D-[GLU]418:D 3.7Å, in average 3.73Å. 310-helix is tighter and bonds are tenser than in usual helixes, so they are longer than common 3Å and angles are smaller than common 180°, 156.1, 132.3 and 157° respectively and 148,5° in average.[3] Hydrogen bonds were found using calculate hbonds command and measurements helped to calculate their parameters.
Stacking
Using restrict aromatic command we identified aminoacids with aromatic radicals in side chains, which can engage in stacking, resulting from interaction of pi-orbitals of conjugated systems. Such interactions are possible if distance between centers of conjugated systems are 4.5-5Å. [4]
|
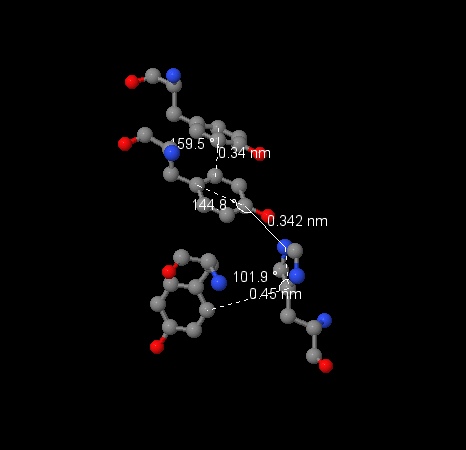
We choosed residues of aromatic aminoacids which seemed most close to each other and measured it to prove that the interaction is possible. We found system of 4 residues: [PHE]-437:D – [PHE]-409:D – [PHE]-453:D – [TYR]-445:D, minimal distances between atoms in their aromatic structures are respectively 3.91Å, 4.67Å и 4.86Å. Angles between ring planes are 141.0° (437 – 409), 128.5° (409 – 453), 135.8° (453 – 445 Another system of 4 residues: [TYR]-171:G – [TYR]-78:G – [HIS]-107:G – [TYR]-113:G, distances between them are 3.4Å, 3.42Å, 4.5Å, which makes the interaction possible. Angles between ring planes are 159.5°, 144.8°, 101.9° (the system can be seen on the image) |
Salt bridges
Such interaction takes place when positively and negatively charged acids engage and is mostly electrostatical. The distance between the residues must be less than 4Å. Aminoacids with positively charged side chains are lysine, arginine and hystidine and negatively charged are aspartat and glutamate. Salt bridges can be found in Jmol by highlighting charged residues and calculating distance between them, but we used online-resource Protein Interaction Calculator. [5]
|
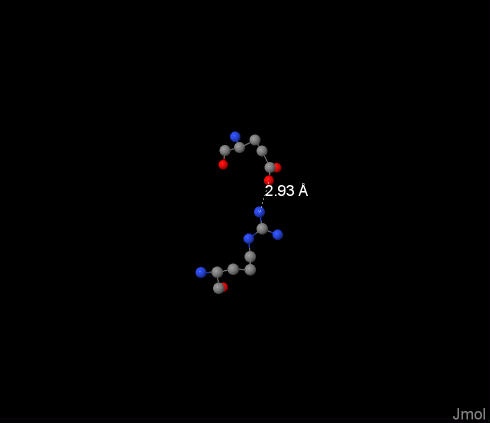
Salt bridge was found between 122:U-Arg и 391:P-Glu, it’s length is 2.93Å, and it can be seen on the image. The distance is small enough for the residues to engage. Salt bridges are essential for structure of the protein, they afflict shape of the molecule and of it’s active site. |
Sulfur-aromatic interaction
Such connections take place between aromatic aminoacids (phenylalanine,thyrosine, thriptophane) and aminoacids with sulfur atom in side chain. It is supposed to be similar to Van-der-Waals interactions since sulfur atom is a great constant dipole. [6] They are essential for protein tertiary structure and connect different sites of aminoacid chain.
|
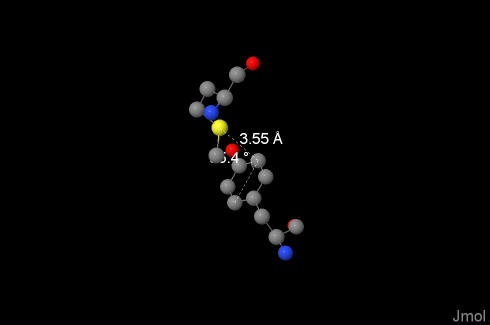
In our molecule they were found using the online-resource, between 199:R-Tyr and 137:N-Met, it is presented on the image, distance between sulfur atom and closest atom of aromatic ring is 3.55Å, angle between the bond and ring plane is 154.23° (calculated online). The engagement is possible if distance is less than 6Å. [7] |
Cation-pi interaction
Positively charged ligands, metal cations or positively charged aminoacid sidechains (lysine, arginine) can interact with negatively charged due to electron delocalization aromatic rings (tyrosine, phenylalanine, tryptophan).
|
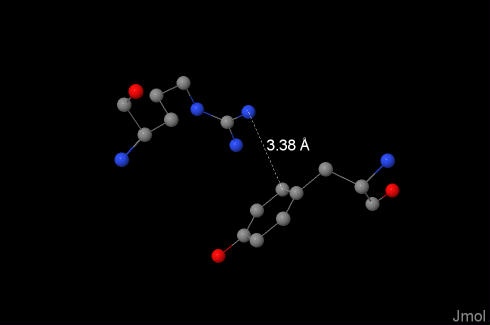
Such bonds explain the protein structure and makes ligand-protein interactions possible. In our molecule they were found between 19:O-Tyr and 171:V-Arg, the online-calculator was used. Distance between positively charged nitrogen atom and the aromatic ring is 3.38Å. |
Jmol-applet showing the interactions
|
|
Load scripts
Sources
3. Lehninger Principles of Biochemistry; Part 1
4. http://www.jbc.org/content/273/25/15458.long
5. http://pic.mbu.iisc.ernet.in
6. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4972207/
7. Sulphur-aromatic interactions in proteins, K.S.C.Reid P.F.Lindley J.M.Thornton
8. Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999 Aug 17;96(17):9459-64. PMID:10449714

