This protein is the part of HIV-1 (Human immunodeficiency virus type 1) capsid. It is t he virus that can lead to acquired immunodeficiency syndrome, or AIDS, if not treated. Unlike some other viruses, the human body can’t get rid of HIV completely, even with treatment. So once you get HIV, you have it for life. HIV-1 displays the unique ability to infect nondividing cells. It is possible throug h the capsid. Nup153 peptide, which is in complex with the protien unlocks the nuclear pore Complex for HIV-1 nuclear translocation in nondividing cells.

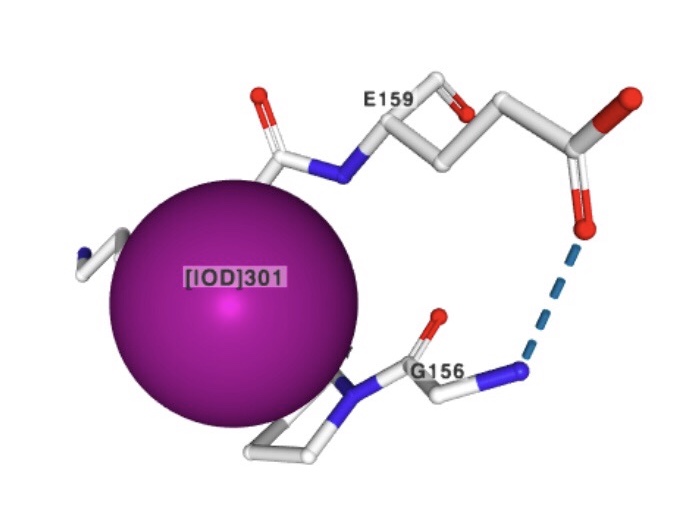
1) Iodide ion
Molecular formula: I-
Molecular weight: 126.904 g/mol
PubChem CID: 30165
Iodide ion can function as an antioxidant as it is a reducing species that can detoxify reactive oxygen species such as hydrogen peroxide.
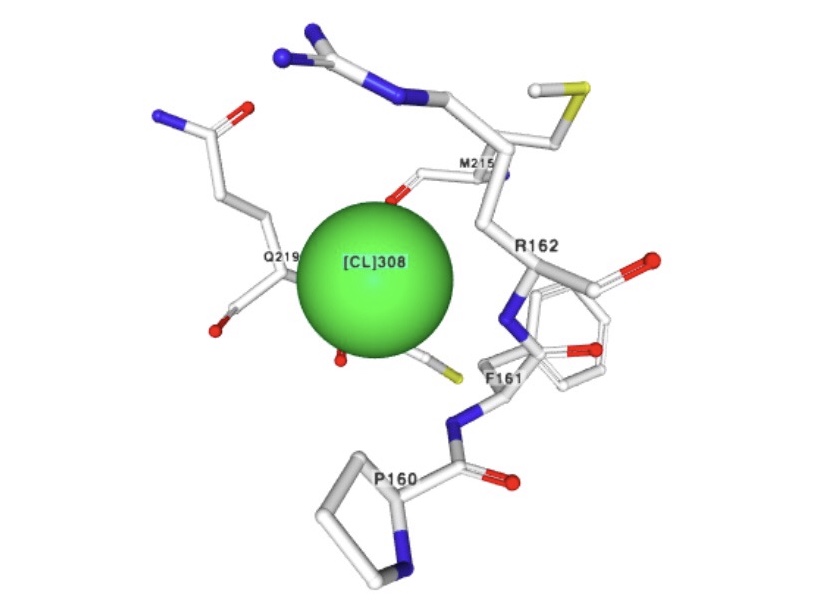
2) Chloride ion
Molecular formula: Cl-
Molecular weight: 35.45 g/mol
PubChem CID: 312
Chloride ions form an inner layer adjacent to the surface of the capsid.
The biological role of this ion is unclear as it be substituting for other ion available in the cytoplasm.
Amide bond is called peptide if it is formed when the carboxyl group of an L-amino acid acylates an amino group of the other amino acid. Peptide bonds in proteins are flat and submitted almost entirely by trans isomers. These bonds are responsible for the primary structure formation. Since any protein has the large number of peptide bonds, here is the image of one of them. The amino acids’ point of contact is expanded.
6aya.jpg)
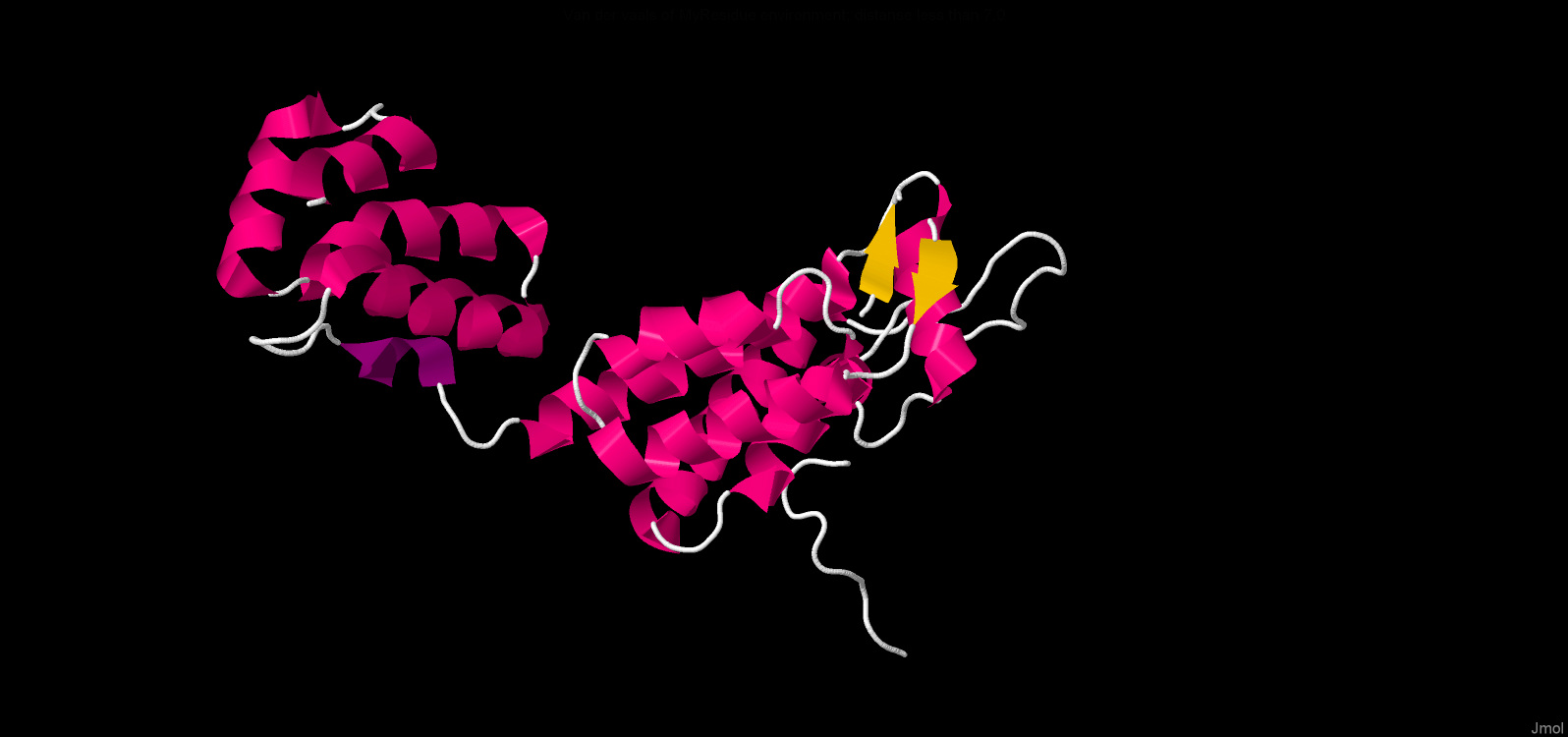
The presense of cysteine residues enables the formation of disulfide bonds which additionally stabilize protein structure. One disulfide bond was found in the ‘6aya’ molecule which is coloured yellow in the picture.
6aya.jpg)
There are alpha helixes and a beta sheet in the secondary structure of this biomolecule (PDB 6aya). Consequently, there are many hydrogen bonds retaining structure. The data below are about hydrogen bonds in our molecule:

| Table 1. Beta-sheets. | |||
|---|---|---|---|
| # | Atom names | Value |
| 1 | O([VAL]3:A - N([VAL]11:A | 2.84 Å |
| 2 | N([VAL]3:A - O([VAL]11:A | 3.00 Å |
| Table 2. Alpha-helix. | |||
|---|---|---|---|
| # | Atom names | Value |
| 1 | O([GLU]212:A - N([THR]216:A | 2.92 Å |
| 2 | O([LEU]211:A - N([MET]215:A | 2.89 Å |
| 3 | O([THR]210:A - N([MET]214:A | 3.09Å |

| Table 3. 3-10 helix. | |||
|---|---|---|---|
| # | Atom names | Value |
| 1 | O([ILE]150:A - N([ILE]153:A | 3.61 Å |
| 2 | O([SER]149:A - N([ASP]152:A | 3.13 Å |
Here are only the lengths of hydrogen bonds, because in the PDB file of the protein 6 aya
there are no protons, it is not possible to measure the angles. However, the data on the
length of hydrogen bonds for each element of the secondary structure coincide with the
theoretical ones. Interestingly, the biomolecule contains not only alpha-helix and
beta-strands, but aslo a 3-10 helix.
Ionogenic groups (glutamic acid, aspartic acid, histidine, lysine and argenine residues) participate in ionic interactions, forming salt bridges. Negatively charged carboxyl radical groups of aspartic and glutamic acids and positively charged radical groups of lysine, argenine, and histidine partcipate in interactions which are possible inside the protein as well as with other molecules. In the 1st picture residues are represented, coloured according to their group’s charge. From the amino acids capable on ionic bonds formation radical groups on distance within 4Å were chosen.
6aya.jpg)
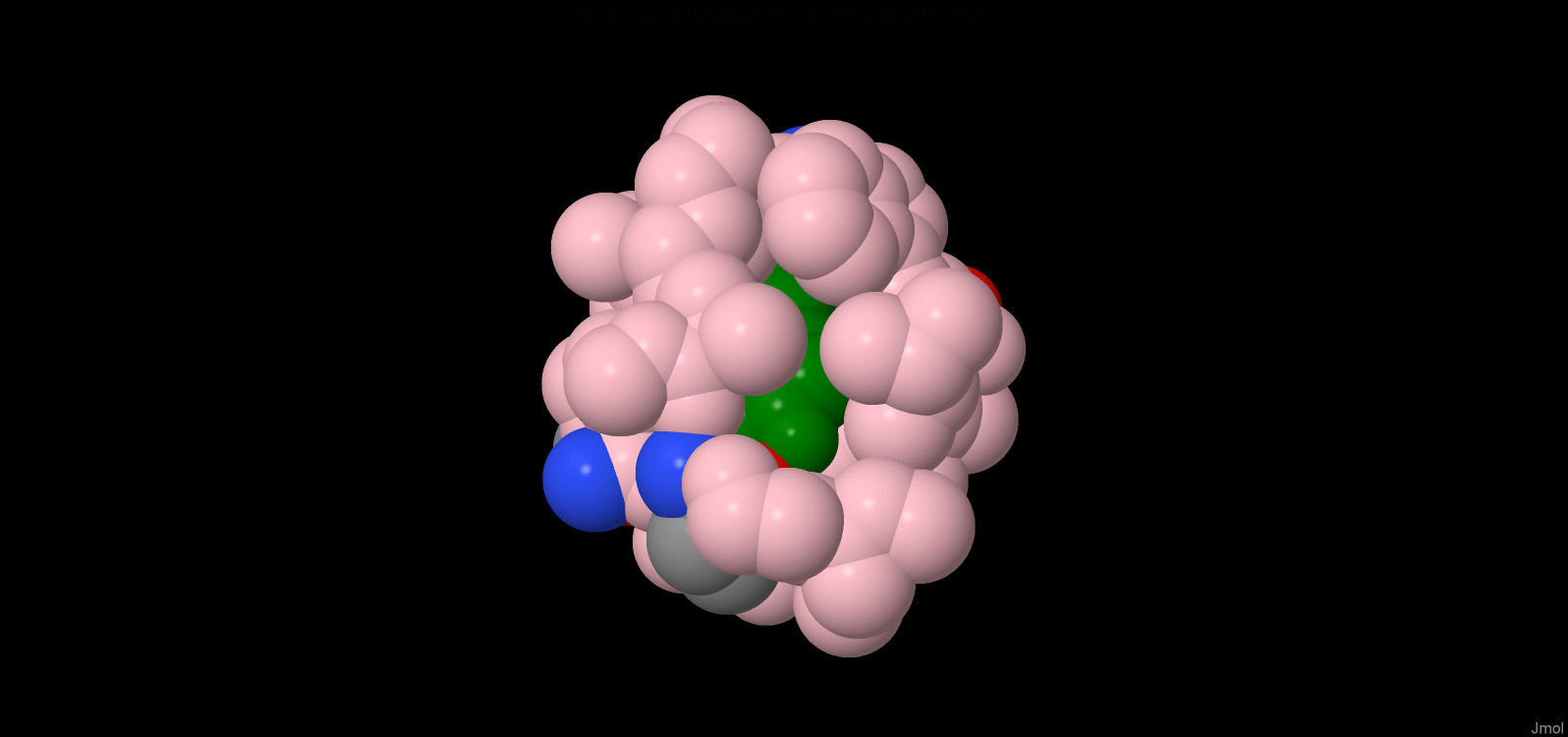
1) Search for contact
In JMOL all hydrophobic residues (which are coloured purple) and Tyr and Phe residues (since they are
quite large - green) were chosen. The residue [Tyr]130:a was then selected because
it best satisfied the search conditions ("a hydrophobic residue whose side chain is
packed in a hydrophobic environment”)
2)Hydrophobic core
The remainder [Tyr] 130: a is almost completely covered by atoms at a distance of no more than 6A. In its hydrophobic shell there is a” hole", which can fit 1 molecule of water.
Distances between neighboring covalently unbound atoms were also measured.Table 4 shows the measurement results.When calculating distances taking into account van der Waals radii, we obtain the following results:
| Table 4. | |||
|---|---|---|---|
| # | Distance between the centers of the atoms, A | Distance considering Van der Waals radii, A | |
| [ILE]134:A.CG2 | [LEU]52:A.CD2 | 6.36 | 2.66 |
| [THR]48:A.C | [GLY]127:A.CA | 4.69 | 0.99 |
| [LEU]52:A.O | [ILE]134:A.CG2 | 6.47 | 3.22 |
| [LEU]52:A.CA | [ILE]134:A.CD1 | 4.74 | 1.04 |
| [LEU]56:A.CD1 | [ILE]73:A.CD1 | 4.21 | 0.51 |
| [ILE]73:A.CG1 | [TRP]133:A.O | 4.94 | 1.69 |
| [VAL]126:A.CB | [ILE]104:A.CA | 4.7 | 1.0 |
| [PRO]49:A.CG | [ILE]104:A.C | 4.33 | 0.64 |
| [ALA]105:A.CB | [PHE]1417:B.CD1 | 7.42 | 3.72 |
| [PHE]1417:B.O | [ILE]73:A.CG2 | 7.13 | 3.88 |
Based on table 4 average distance between atoms not bound covalently was calculated = 1.93A so water molecule can't fit. As has been said in the 1st paragraph, there's only one place in hydrophobic shell where distance between atoms is enough for water molecule to fit there (we assume that the water molecule is approximately equal to the oxygen atom). 7.13-(1.85+1.4)=3.88А and 7.42-(1.85+1.85)=3.72А. The diameter of the oxygen atom is assumed to be 2.8A. The H2O molecule is fit indeed.